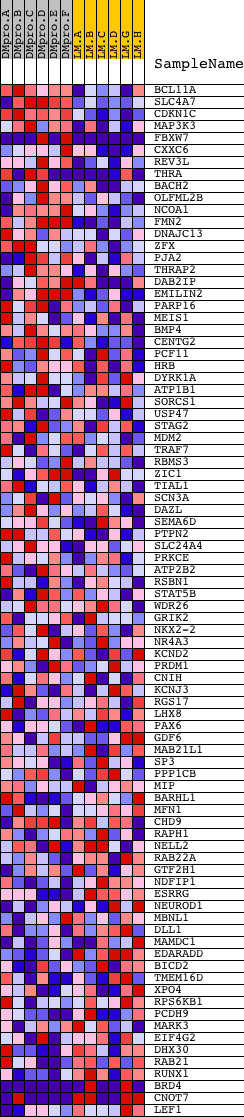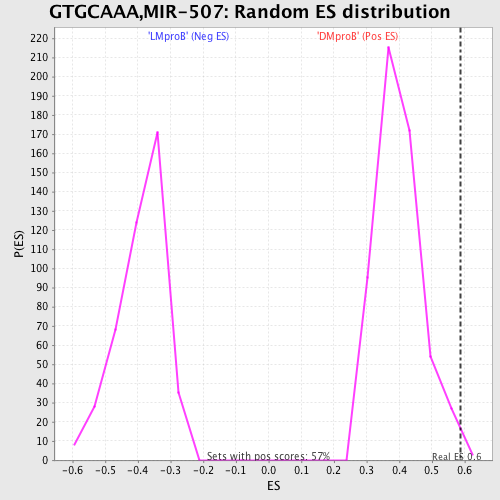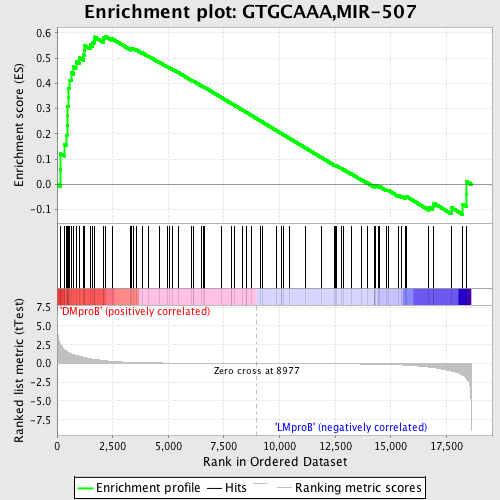
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | DMproB |
| GeneSet | GTGCAAA,MIR-507 |
| Enrichment Score (ES) | 0.5867332 |
| Normalized Enrichment Score (NES) | 1.4745351 |
| Nominal p-value | 0.0053003533 |
| FDR q-value | 0.52565837 |
| FWER p-Value | 0.951 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BCL11A | 4691 | 147 | 2.605 | 0.0574 | Yes | ||
| 2 | SLC4A7 | 10181 | 160 | 2.535 | 0.1204 | Yes | ||
| 3 | CDKN1C | 17546 | 315 | 1.898 | 0.1597 | Yes | ||
| 4 | MAP3K3 | 20626 | 424 | 1.629 | 0.1947 | Yes | ||
| 5 | FBXW7 | 1805 11928 | 450 | 1.581 | 0.2330 | Yes | ||
| 6 | CXXC6 | 19747 | 472 | 1.553 | 0.2708 | Yes | ||
| 7 | REV3L | 20050 | 478 | 1.541 | 0.3092 | Yes | ||
| 8 | THRA | 1447 10171 1406 | 513 | 1.480 | 0.3445 | Yes | ||
| 9 | BACH2 | 8648 | 521 | 1.471 | 0.3810 | Yes | ||
| 10 | OLFML2B | 11496 | 573 | 1.375 | 0.4127 | Yes | ||
| 11 | NCOA1 | 5154 | 636 | 1.281 | 0.4415 | Yes | ||
| 12 | FMN2 | 14037 | 742 | 1.155 | 0.4648 | Yes | ||
| 13 | DNAJC13 | 6177 | 850 | 1.054 | 0.4855 | Yes | ||
| 14 | ZFX | 5984 | 1002 | 0.958 | 0.5014 | Yes | ||
| 15 | PJA2 | 5904 | 1186 | 0.821 | 0.5121 | Yes | ||
| 16 | THRAP2 | 8009 | 1224 | 0.796 | 0.5301 | Yes | ||
| 17 | DAB2IP | 12808 2692 | 1237 | 0.786 | 0.5491 | Yes | ||
| 18 | EMILIN2 | 10901 | 1485 | 0.635 | 0.5517 | Yes | ||
| 19 | PARP16 | 19405 | 1604 | 0.579 | 0.5599 | Yes | ||
| 20 | MEIS1 | 20524 | 1660 | 0.553 | 0.5708 | Yes | ||
| 21 | BMP4 | 21863 | 1682 | 0.544 | 0.5833 | Yes | ||
| 22 | CENTG2 | 6874 | 2067 | 0.392 | 0.5724 | Yes | ||
| 23 | PCF11 | 121 | 2087 | 0.385 | 0.5810 | Yes | ||
| 24 | HRB | 14208 4066 | 2153 | 0.366 | 0.5867 | Yes | ||
| 25 | DYRK1A | 4649 | 2488 | 0.278 | 0.5757 | No | ||
| 26 | ATP1B1 | 4420 | 3304 | 0.150 | 0.5355 | No | ||
| 27 | SORCS1 | 7184 12203 | 3343 | 0.146 | 0.5371 | No | ||
| 28 | USP47 | 6794 | 3419 | 0.139 | 0.5365 | No | ||
| 29 | STAG2 | 5521 | 3550 | 0.126 | 0.5327 | No | ||
| 30 | MDM2 | 19620 3327 | 3820 | 0.107 | 0.5208 | No | ||
| 31 | TRAF7 | 10325 | 4118 | 0.090 | 0.5071 | No | ||
| 32 | RBMS3 | 18973 | 4601 | 0.070 | 0.4828 | No | ||
| 33 | ZIC1 | 19040 | 4972 | 0.059 | 0.4643 | No | ||
| 34 | TIAL1 | 996 5753 | 5066 | 0.057 | 0.4607 | No | ||
| 35 | SCN3A | 14573 | 5175 | 0.054 | 0.4563 | No | ||
| 36 | DAZL | 22944 | 5463 | 0.047 | 0.4420 | No | ||
| 37 | SEMA6D | 2800 14876 90 | 6045 | 0.036 | 0.4115 | No | ||
| 38 | PTPN2 | 23414 | 6121 | 0.035 | 0.4084 | No | ||
| 39 | SLC24A4 | 6223 | 6146 | 0.035 | 0.4079 | No | ||
| 40 | PRKCE | 9575 | 6468 | 0.029 | 0.3914 | No | ||
| 41 | ATP2B2 | 17038 | 6573 | 0.028 | 0.3864 | No | ||
| 42 | RSBN1 | 6035 | 6614 | 0.027 | 0.3850 | No | ||
| 43 | STAT5B | 20222 | 7378 | 0.018 | 0.3442 | No | ||
| 44 | WDR26 | 13733 | 7820 | 0.013 | 0.3208 | No | ||
| 45 | GRIK2 | 3318 | 7837 | 0.012 | 0.3202 | No | ||
| 46 | NKX2-2 | 5176 | 7969 | 0.011 | 0.3134 | No | ||
| 47 | NR4A3 | 9473 16212 5183 | 8349 | 0.006 | 0.2931 | No | ||
| 48 | KCND2 | 4941 17515 | 8517 | 0.005 | 0.2842 | No | ||
| 49 | PRDM1 | 19775 3337 | 8527 | 0.004 | 0.2838 | No | ||
| 50 | CNIH | 4536 23971 | 8715 | 0.003 | 0.2738 | No | ||
| 51 | KCNJ3 | 4944 | 9162 | -0.002 | 0.2498 | No | ||
| 52 | RGS17 | 7133 | 9214 | -0.003 | 0.2471 | No | ||
| 53 | LHX8 | 4997 | 9859 | -0.008 | 0.2126 | No | ||
| 54 | PAX6 | 5223 | 10074 | -0.011 | 0.2013 | No | ||
| 55 | GDF6 | 10814 | 10077 | -0.011 | 0.2014 | No | ||
| 56 | MAB21L1 | 15589 5046 | 10195 | -0.012 | 0.1954 | No | ||
| 57 | SP3 | 5483 | 10452 | -0.015 | 0.1820 | No | ||
| 58 | PPP1CB | 5282 3529 3504 | 11155 | -0.023 | 0.1447 | No | ||
| 59 | MIP | 19845 | 11886 | -0.032 | 0.1061 | No | ||
| 60 | BARHL1 | 14634 | 12456 | -0.041 | 0.0764 | No | ||
| 61 | MFN1 | 12529 1823 | 12526 | -0.042 | 0.0737 | No | ||
| 62 | CHD9 | 6784 11553 18531 | 12541 | -0.042 | 0.0741 | No | ||
| 63 | RAPH1 | 19110 | 12788 | -0.047 | 0.0620 | No | ||
| 64 | NELL2 | 12006 7020 22151 | 12894 | -0.049 | 0.0575 | No | ||
| 65 | RAB22A | 2888 5343 | 13210 | -0.056 | 0.0419 | No | ||
| 66 | GTF2H1 | 4069 18236 | 13701 | -0.070 | 0.0172 | No | ||
| 67 | NDFIP1 | 23573 | 13932 | -0.079 | 0.0068 | No | ||
| 68 | ESRRG | 6447 | 14264 | -0.092 | -0.0088 | No | ||
| 69 | NEUROD1 | 14550 | 14275 | -0.092 | -0.0070 | No | ||
| 70 | MBNL1 | 1921 15582 | 14280 | -0.093 | -0.0049 | No | ||
| 71 | DLL1 | 23119 | 14314 | -0.094 | -0.0043 | No | ||
| 72 | MAMDC1 | 21054 6798 | 14448 | -0.101 | -0.0090 | No | ||
| 73 | EDARADD | 5056 3282 3278 | 14511 | -0.104 | -0.0097 | No | ||
| 74 | BICD2 | 8048 | 14809 | -0.121 | -0.0227 | No | ||
| 75 | TMEM16D | 19649 | 14873 | -0.125 | -0.0229 | No | ||
| 76 | XPO4 | 7161 | 15327 | -0.166 | -0.0432 | No | ||
| 77 | RPS6KB1 | 7815 1207 13040 | 15488 | -0.186 | -0.0472 | No | ||
| 78 | PCDH9 | 21736 | 15662 | -0.211 | -0.0512 | No | ||
| 79 | MARK3 | 9369 | 15686 | -0.215 | -0.0471 | No | ||
| 80 | EIF4G2 | 1908 8892 | 16710 | -0.470 | -0.0905 | No | ||
| 81 | DHX30 | 18989 | 16895 | -0.550 | -0.0866 | No | ||
| 82 | RAB21 | 5696 | 16937 | -0.564 | -0.0747 | No | ||
| 83 | RUNX1 | 4481 | 17719 | -1.000 | -0.0917 | No | ||
| 84 | BRD4 | 7164 1609 1628 1598 1552 1601 12176 1630 1501 | 18231 | -1.573 | -0.0799 | No | ||
| 85 | CNOT7 | 3787 9600 | 18398 | -1.966 | -0.0395 | No | ||
| 86 | LEF1 | 1860 15420 | 18411 | -2.043 | 0.0111 | No |