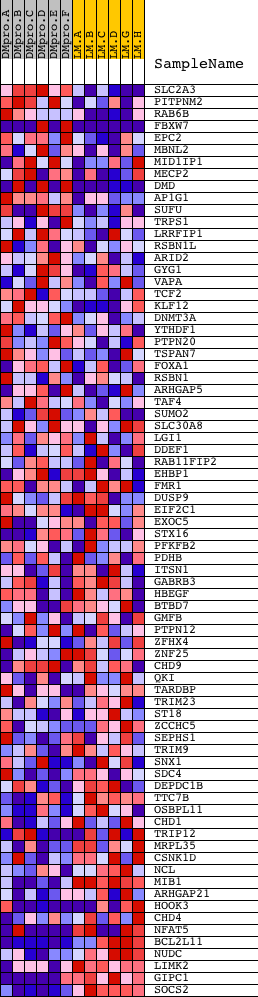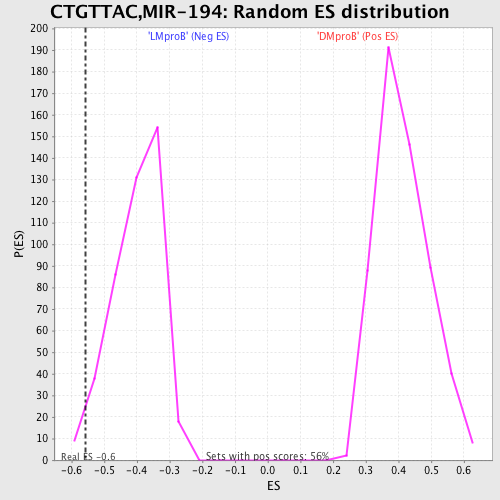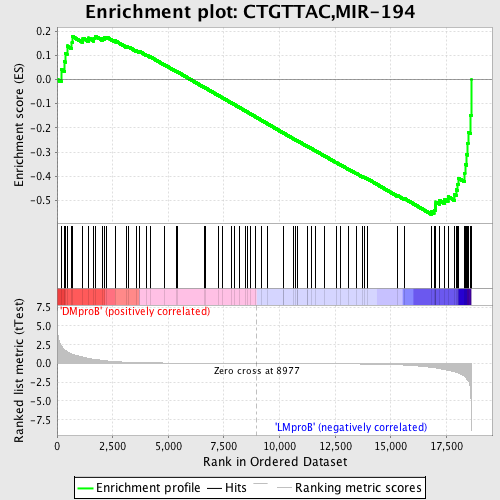
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | CTGTTAC,MIR-194 |
| Enrichment Score (ES) | -0.55797577 |
| Normalized Enrichment Score (NES) | -1.4012088 |
| Nominal p-value | 0.025229357 |
| FDR q-value | 0.6595679 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SLC2A3 | 17010 | 183 | 2.386 | 0.0413 | No | ||
| 2 | PITPNM2 | 16371 | 326 | 1.867 | 0.0737 | No | ||
| 3 | RAB6B | 11289 | 366 | 1.742 | 0.1089 | No | ||
| 4 | FBXW7 | 1805 11928 | 450 | 1.581 | 0.1384 | No | ||
| 5 | EPC2 | 15015 | 668 | 1.235 | 0.1531 | No | ||
| 6 | MBNL2 | 21932 | 702 | 1.192 | 0.1769 | No | ||
| 7 | MID1IP1 | 24381 | 1160 | 0.841 | 0.1703 | No | ||
| 8 | MECP2 | 5088 | 1412 | 0.677 | 0.1713 | No | ||
| 9 | DMD | 24295 2647 | 1652 | 0.559 | 0.1704 | No | ||
| 10 | AP1G1 | 4392 | 1706 | 0.534 | 0.1790 | No | ||
| 11 | SUFU | 6283 | 2040 | 0.402 | 0.1697 | No | ||
| 12 | TRPS1 | 8195 | 2140 | 0.370 | 0.1723 | No | ||
| 13 | LRRFIP1 | 14191 4007 3965 | 2236 | 0.344 | 0.1745 | No | ||
| 14 | RSBN1L | 16598 | 2608 | 0.250 | 0.1599 | No | ||
| 15 | ARID2 | 8062 13378 | 3125 | 0.171 | 0.1357 | No | ||
| 16 | GYG1 | 1820 15371 | 3226 | 0.158 | 0.1337 | No | ||
| 17 | VAPA | 22908 | 3566 | 0.124 | 0.1181 | No | ||
| 18 | TCF2 | 1395 20727 | 3688 | 0.116 | 0.1141 | No | ||
| 19 | KLF12 | 4960 2637 21732 9228 | 3693 | 0.116 | 0.1163 | No | ||
| 20 | DNMT3A | 2167 21330 | 4026 | 0.094 | 0.1004 | No | ||
| 21 | YTHDF1 | 14311 | 4182 | 0.087 | 0.0939 | No | ||
| 22 | PTPN20 | 22049 | 4831 | 0.063 | 0.0603 | No | ||
| 23 | TSPAN7 | 24382 | 5376 | 0.049 | 0.0320 | No | ||
| 24 | FOXA1 | 21060 | 5410 | 0.049 | 0.0313 | No | ||
| 25 | RSBN1 | 6035 | 6614 | 0.027 | -0.0330 | No | ||
| 26 | ARHGAP5 | 4412 8625 | 6658 | 0.027 | -0.0347 | No | ||
| 27 | TAF4 | 14319 | 7240 | 0.019 | -0.0657 | No | ||
| 28 | SUMO2 | 1293 9319 | 7412 | 0.017 | -0.0745 | No | ||
| 29 | SLC30A8 | 10737 22478 | 7839 | 0.012 | -0.0972 | No | ||
| 30 | LGI1 | 23867 | 7971 | 0.011 | -0.1041 | No | ||
| 31 | DDEF1 | 22282 2253 | 8218 | 0.008 | -0.1172 | No | ||
| 32 | RAB11FIP2 | 23635 | 8463 | 0.005 | -0.1302 | No | ||
| 33 | EHBP1 | 20516 | 8574 | 0.004 | -0.1361 | No | ||
| 34 | FMR1 | 24321 | 8683 | 0.003 | -0.1418 | No | ||
| 35 | DUSP9 | 24307 | 8900 | 0.001 | -0.1535 | No | ||
| 36 | EIF2C1 | 10672 | 9174 | -0.002 | -0.1681 | No | ||
| 37 | EXOC5 | 8370 | 9466 | -0.005 | -0.1837 | No | ||
| 38 | STX16 | 10464 | 10181 | -0.012 | -0.2220 | No | ||
| 39 | PFKFB2 | 4116 5242 | 10196 | -0.012 | -0.2225 | No | ||
| 40 | PDHB | 12670 7548 | 10624 | -0.017 | -0.2452 | No | ||
| 41 | ITSN1 | 9196 | 10712 | -0.018 | -0.2495 | No | ||
| 42 | GABRB3 | 4747 | 10801 | -0.019 | -0.2538 | No | ||
| 43 | HBEGF | 23457 | 10808 | -0.019 | -0.2537 | No | ||
| 44 | BTBD7 | 20998 | 11234 | -0.024 | -0.2761 | No | ||
| 45 | GMFB | 21861 | 11274 | -0.024 | -0.2777 | No | ||
| 46 | PTPN12 | 16597 3502 3586 3665 | 11449 | -0.026 | -0.2865 | No | ||
| 47 | ZFHX4 | 8158 | 11619 | -0.029 | -0.2950 | No | ||
| 48 | ZNF25 | 17025 | 12004 | -0.034 | -0.3150 | No | ||
| 49 | CHD9 | 6784 11553 18531 | 12541 | -0.042 | -0.3430 | No | ||
| 50 | QKI | 23128 5340 | 12735 | -0.046 | -0.3525 | No | ||
| 51 | TARDBP | 10520 6066 | 13099 | -0.053 | -0.3709 | No | ||
| 52 | TRIM23 | 8165 3230 | 13453 | -0.062 | -0.3886 | No | ||
| 53 | ST18 | 14298 | 13731 | -0.071 | -0.4020 | No | ||
| 54 | ZCCHC5 | 24081 | 13802 | -0.074 | -0.4042 | No | ||
| 55 | SEPHS1 | 15125 | 13966 | -0.080 | -0.4113 | No | ||
| 56 | TRIM9 | 2118 8076 13577 8234 | 15299 | -0.163 | -0.4796 | No | ||
| 57 | SNX1 | 19076 | 15623 | -0.206 | -0.4926 | No | ||
| 58 | SDC4 | 14349 | 16836 | -0.530 | -0.5466 | Yes | ||
| 59 | DEPDC1B | 5757 | 16942 | -0.566 | -0.5401 | Yes | ||
| 60 | TTC7B | 21006 | 16998 | -0.586 | -0.5305 | Yes | ||
| 61 | OSBPL11 | 22781 | 17012 | -0.594 | -0.5185 | Yes | ||
| 62 | CHD1 | 23377 | 17027 | -0.600 | -0.5064 | Yes | ||
| 63 | TRIP12 | 4813 9055 9054 | 17194 | -0.676 | -0.5008 | Yes | ||
| 64 | MRPL35 | 7267 | 17415 | -0.812 | -0.4953 | Yes | ||
| 65 | CSNK1D | 4181 8352 1387 | 17573 | -0.903 | -0.4844 | Yes | ||
| 66 | NCL | 5153 13899 | 17866 | -1.091 | -0.4767 | Yes | ||
| 67 | MIB1 | 5908 | 17949 | -1.166 | -0.4561 | Yes | ||
| 68 | ARHGAP21 | 12929 | 17990 | -1.197 | -0.4326 | Yes | ||
| 69 | HOOK3 | 18903 6734 | 18031 | -1.247 | -0.4080 | Yes | ||
| 70 | CHD4 | 8418 17281 4225 | 18312 | -1.692 | -0.3868 | Yes | ||
| 71 | NFAT5 | 3921 7037 12036 | 18335 | -1.759 | -0.3503 | Yes | ||
| 72 | BCL2L11 | 2790 14861 | 18415 | -2.056 | -0.3105 | Yes | ||
| 73 | NUDC | 9495 | 18454 | -2.205 | -0.2652 | Yes | ||
| 74 | LIMK2 | 9278 1296 | 18473 | -2.247 | -0.2180 | Yes | ||
| 75 | GIPC1 | 3841 3882 18552 | 18587 | -3.539 | -0.1482 | Yes | ||
| 76 | SOCS2 | 5694 | 18613 | -6.981 | 0.0002 | Yes |