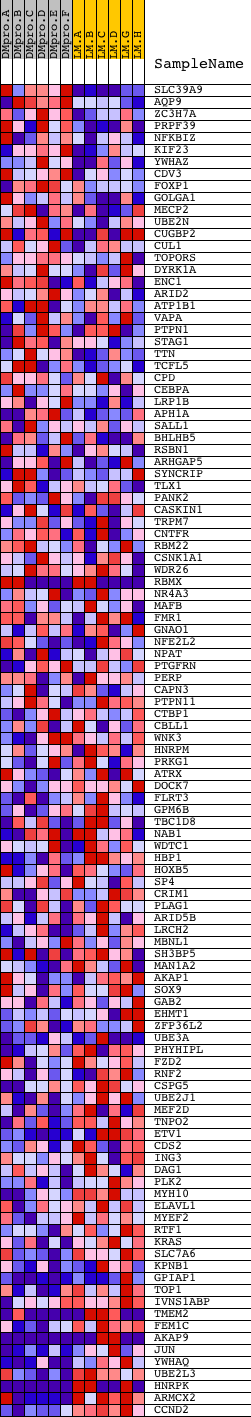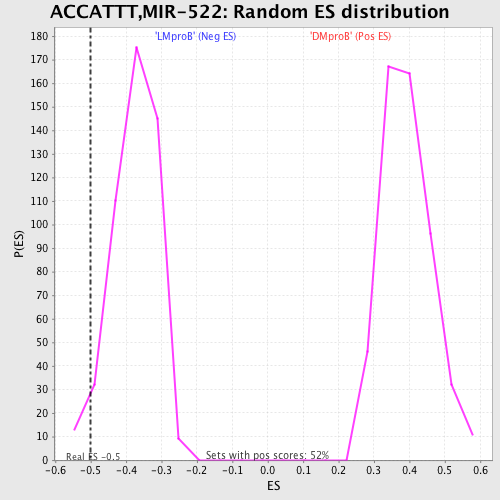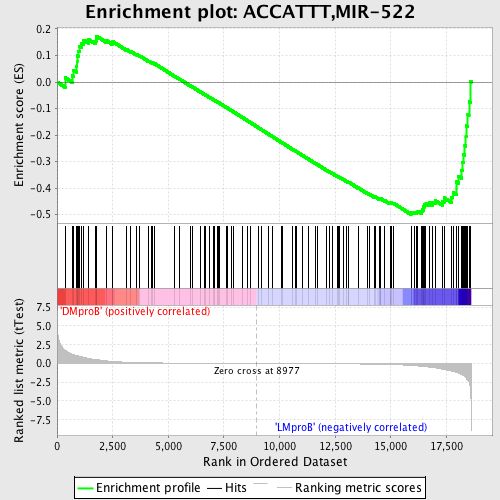
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | ACCATTT,MIR-522 |
| Enrichment Score (ES) | -0.4991605 |
| Normalized Enrichment Score (NES) | -1.3248543 |
| Nominal p-value | 0.04338843 |
| FDR q-value | 0.65454185 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SLC39A9 | 11597 | 360 | 1.747 | 0.0164 | No | ||
| 2 | AQP9 | 7218 | 687 | 1.209 | 0.0236 | No | ||
| 3 | ZC3H7A | 22663 | 750 | 1.143 | 0.0438 | No | ||
| 4 | PRPF39 | 21259 | 867 | 1.039 | 0.0588 | No | ||
| 5 | NFKBIZ | 22575 | 893 | 1.026 | 0.0786 | No | ||
| 6 | KIF23 | 19091 | 897 | 1.024 | 0.0994 | No | ||
| 7 | YWHAZ | 10370 | 970 | 0.984 | 0.1158 | No | ||
| 8 | CDV3 | 11587 | 1006 | 0.957 | 0.1335 | No | ||
| 9 | FOXP1 | 4242 | 1101 | 0.886 | 0.1466 | No | ||
| 10 | GOLGA1 | 14595 | 1195 | 0.816 | 0.1584 | No | ||
| 11 | MECP2 | 5088 | 1412 | 0.677 | 0.1606 | No | ||
| 12 | UBE2N | 8216 19898 | 1723 | 0.529 | 0.1547 | No | ||
| 13 | CUGBP2 | 4687 8923 | 1751 | 0.515 | 0.1638 | No | ||
| 14 | CUL1 | 17462 | 1773 | 0.507 | 0.1731 | No | ||
| 15 | TOPORS | 15920 | 2199 | 0.353 | 0.1574 | No | ||
| 16 | DYRK1A | 4649 | 2488 | 0.278 | 0.1475 | No | ||
| 17 | ENC1 | 21579 | 2502 | 0.274 | 0.1525 | No | ||
| 18 | ARID2 | 8062 13378 | 3125 | 0.171 | 0.1224 | No | ||
| 19 | ATP1B1 | 4420 | 3304 | 0.150 | 0.1159 | No | ||
| 20 | VAPA | 22908 | 3566 | 0.124 | 0.1043 | No | ||
| 21 | PTPN1 | 5325 | 3722 | 0.113 | 0.0983 | No | ||
| 22 | STAG1 | 5520 9905 2987 | 4115 | 0.090 | 0.0789 | No | ||
| 23 | TTN | 14552 2743 14553 | 4250 | 0.083 | 0.0734 | No | ||
| 24 | TCFL5 | 14313 | 4284 | 0.082 | 0.0733 | No | ||
| 25 | CPD | 20344 4556 | 4395 | 0.078 | 0.0690 | No | ||
| 26 | CEBPA | 4514 | 5270 | 0.052 | 0.0228 | No | ||
| 27 | LRP1B | 8249 | 5521 | 0.046 | 0.0102 | No | ||
| 28 | APH1A | 15501 5947 10378 | 5977 | 0.038 | -0.0136 | No | ||
| 29 | SALL1 | 18796 12207 | 6090 | 0.036 | -0.0189 | No | ||
| 30 | BHLHB5 | 15629 | 6433 | 0.030 | -0.0368 | No | ||
| 31 | RSBN1 | 6035 | 6614 | 0.027 | -0.0459 | No | ||
| 32 | ARHGAP5 | 4412 8625 | 6658 | 0.027 | -0.0477 | No | ||
| 33 | SYNCRIP | 3078 3035 3107 | 6830 | 0.024 | -0.0565 | No | ||
| 34 | TLX1 | 23839 | 7021 | 0.022 | -0.0663 | No | ||
| 35 | PANK2 | 14838 | 7067 | 0.021 | -0.0683 | No | ||
| 36 | CASKIN1 | 11219 879 | 7217 | 0.020 | -0.0759 | No | ||
| 37 | TRPM7 | 14446 | 7247 | 0.019 | -0.0771 | No | ||
| 38 | CNTFR | 2515 15906 | 7278 | 0.019 | -0.0783 | No | ||
| 39 | RBM22 | 23542 | 7633 | 0.015 | -0.0972 | No | ||
| 40 | CSNK1A1 | 8204 | 7661 | 0.014 | -0.0983 | No | ||
| 41 | WDR26 | 13733 | 7820 | 0.013 | -0.1066 | No | ||
| 42 | RBMX | 5367 9708 | 7931 | 0.011 | -0.1123 | No | ||
| 43 | NR4A3 | 9473 16212 5183 | 8349 | 0.006 | -0.1347 | No | ||
| 44 | MAFB | 14367 | 8570 | 0.004 | -0.1465 | No | ||
| 45 | FMR1 | 24321 | 8683 | 0.003 | -0.1525 | No | ||
| 46 | GNAO1 | 4785 3829 | 9038 | -0.001 | -0.1716 | No | ||
| 47 | NFE2L2 | 2898 14557 | 9171 | -0.002 | -0.1787 | No | ||
| 48 | NPAT | 19452 6356 | 9507 | -0.005 | -0.1967 | No | ||
| 49 | PTGFRN | 15224 | 9694 | -0.007 | -0.2066 | No | ||
| 50 | PERP | 2259 22121 20085 | 10068 | -0.011 | -0.2265 | No | ||
| 51 | CAPN3 | 8686 2868 | 10145 | -0.011 | -0.2304 | No | ||
| 52 | PTPN11 | 5326 16391 9660 | 10601 | -0.016 | -0.2547 | No | ||
| 53 | CTBP1 | 16566 | 10713 | -0.018 | -0.2603 | No | ||
| 54 | CBLL1 | 4188 | 10756 | -0.018 | -0.2622 | No | ||
| 55 | WNK3 | 24227 | 11011 | -0.021 | -0.2755 | No | ||
| 56 | HNRPM | 1511 13370 | 11279 | -0.024 | -0.2894 | No | ||
| 57 | PRKG1 | 5289 | 11622 | -0.029 | -0.3073 | No | ||
| 58 | ATRX | 10350 5922 2628 | 11722 | -0.030 | -0.3120 | No | ||
| 59 | DOCK7 | 12506 | 12099 | -0.035 | -0.3316 | No | ||
| 60 | FLRT3 | 7730 12930 2673 | 12262 | -0.038 | -0.3396 | No | ||
| 61 | GPM6B | 9034 | 12361 | -0.040 | -0.3441 | No | ||
| 62 | TBC1D8 | 12044 | 12609 | -0.044 | -0.3565 | No | ||
| 63 | NAB1 | 9442 9443 5145 5144 | 12650 | -0.044 | -0.3578 | No | ||
| 64 | WDTC1 | 10512 | 12696 | -0.045 | -0.3593 | No | ||
| 65 | HBP1 | 2048 2162 21086 | 12861 | -0.048 | -0.3672 | No | ||
| 66 | HOXB5 | 20687 | 12998 | -0.051 | -0.3735 | No | ||
| 67 | SP4 | 20972 | 13095 | -0.053 | -0.3776 | No | ||
| 68 | CRIM1 | 403 | 13113 | -0.053 | -0.3774 | No | ||
| 69 | PLAG1 | 7148 15949 12161 | 13539 | -0.065 | -0.3990 | No | ||
| 70 | ARID5B | 7728 | 13955 | -0.080 | -0.4198 | No | ||
| 71 | LRCH2 | 24035 | 14032 | -0.083 | -0.4222 | No | ||
| 72 | MBNL1 | 1921 15582 | 14280 | -0.093 | -0.4337 | No | ||
| 73 | SH3BP5 | 6278 | 14288 | -0.093 | -0.4322 | No | ||
| 74 | MAN1A2 | 5076 5075 | 14505 | -0.104 | -0.4417 | No | ||
| 75 | AKAP1 | 4364 1326 1286 20299 | 14518 | -0.104 | -0.4402 | No | ||
| 76 | SOX9 | 20611 | 14554 | -0.106 | -0.4399 | No | ||
| 77 | GAB2 | 1821 18184 2025 | 14724 | -0.116 | -0.4467 | No | ||
| 78 | EHMT1 | 13401 14670 2756 8090 | 14963 | -0.133 | -0.4568 | No | ||
| 79 | ZFP36L2 | 8662 | 15006 | -0.136 | -0.4563 | No | ||
| 80 | UBE3A | 1209 1084 1830 | 15011 | -0.136 | -0.4537 | No | ||
| 81 | PHYHIPL | 12907 7711 | 15118 | -0.146 | -0.4564 | No | ||
| 82 | FZD2 | 12179 | 15910 | -0.252 | -0.4940 | Yes | ||
| 83 | RNF2 | 9728 | 15940 | -0.259 | -0.4902 | Yes | ||
| 84 | CSPG5 | 19296 | 16042 | -0.282 | -0.4899 | Yes | ||
| 85 | UBE2J1 | 16251 | 16158 | -0.308 | -0.4898 | Yes | ||
| 86 | MEF2D | 9379 | 16214 | -0.323 | -0.4861 | Yes | ||
| 87 | TNPO2 | 5608 | 16372 | -0.363 | -0.4872 | Yes | ||
| 88 | ETV1 | 4688 8925 8924 | 16408 | -0.374 | -0.4814 | Yes | ||
| 89 | CDS2 | 14837 | 16478 | -0.391 | -0.4771 | Yes | ||
| 90 | ING3 | 17514 1019 | 16481 | -0.391 | -0.4691 | Yes | ||
| 91 | DAG1 | 18996 8837 | 16519 | -0.402 | -0.4629 | Yes | ||
| 92 | PLK2 | 21566 | 16579 | -0.421 | -0.4574 | Yes | ||
| 93 | MYH10 | 8077 | 16724 | -0.474 | -0.4555 | Yes | ||
| 94 | ELAVL1 | 4888 | 16882 | -0.547 | -0.4527 | Yes | ||
| 95 | MYEF2 | 5140 | 16992 | -0.585 | -0.4466 | Yes | ||
| 96 | RTF1 | 14899 | 17332 | -0.759 | -0.4493 | Yes | ||
| 97 | KRAS | 9247 | 17394 | -0.797 | -0.4363 | Yes | ||
| 98 | SLC7A6 | 18483 | 17737 | -1.008 | -0.4340 | Yes | ||
| 99 | KPNB1 | 20274 | 17794 | -1.043 | -0.4157 | Yes | ||
| 100 | GPIAP1 | 2819 7014 | 17945 | -1.162 | -0.3999 | Yes | ||
| 101 | TOP1 | 5790 5789 10210 | 17946 | -1.163 | -0.3760 | Yes | ||
| 102 | IVNS1ABP | 4384 4050 | 18039 | -1.265 | -0.3550 | Yes | ||
| 103 | TMEM2 | 13517 8193 | 18176 | -1.471 | -0.3322 | Yes | ||
| 104 | FEM1C | 23441 10771 23442 | 18209 | -1.542 | -0.3022 | Yes | ||
| 105 | AKAP9 | 16604 16603 4150 8303 | 18253 | -1.590 | -0.2719 | Yes | ||
| 106 | JUN | 15832 | 18330 | -1.748 | -0.2401 | Yes | ||
| 107 | YWHAQ | 10369 | 18378 | -1.909 | -0.2034 | Yes | ||
| 108 | UBE2L3 | 5818 22647 | 18386 | -1.929 | -0.1642 | Yes | ||
| 109 | HNRPK | 9106 9105 3265 4861 | 18464 | -2.232 | -0.1225 | Yes | ||
| 110 | ARMCX2 | 12530 | 18515 | -2.511 | -0.0736 | Yes | ||
| 111 | CCND2 | 16987 | 18590 | -3.849 | 0.0014 | Yes |