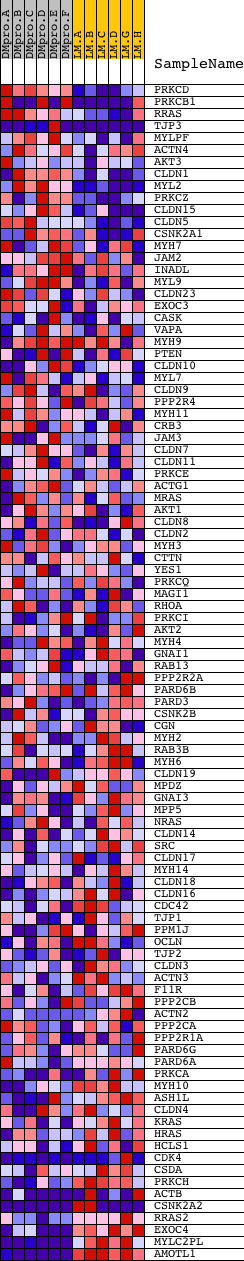
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | HSA04530_TIGHT_JUNCTION |
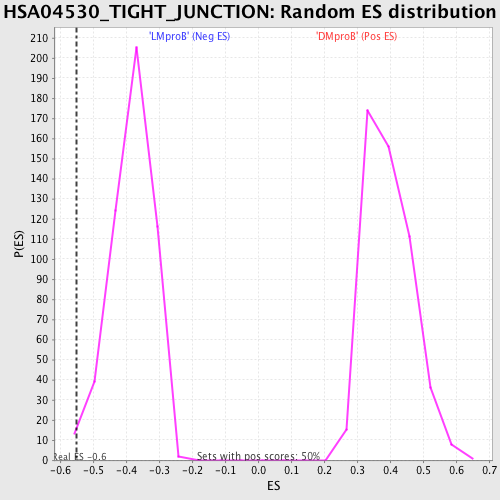
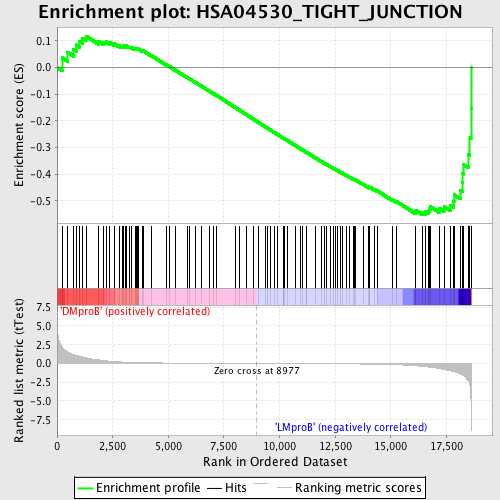
| Enrichment Score (ES) | -0.5505098 |
| Normalized Enrichment Score (NES) | -1.433801 |
| Nominal p-value | 0.01002004 |
| FDR q-value | 0.80578804 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PRKCD | 21897 | 229 | 2.169 | 0.0357 | No | ||
| 2 | PRKCB1 | 1693 9574 | 479 | 1.540 | 0.0564 | No | ||
| 3 | RRAS | 18256 | 739 | 1.158 | 0.0681 | No | ||
| 4 | TJP3 | 19677 3298 | 868 | 1.039 | 0.0842 | No | ||
| 5 | MYLPF | 18069 | 1021 | 0.944 | 0.0970 | No | ||
| 6 | ACTN4 | 17905 1798 1983 | 1141 | 0.856 | 0.1095 | No | ||
| 7 | AKT3 | 13739 982 | 1301 | 0.740 | 0.1173 | No | ||
| 8 | CLDN1 | 22623 | 1867 | 0.471 | 0.0973 | No | ||
| 9 | MYL2 | 16713 | 2079 | 0.387 | 0.0945 | No | ||
| 10 | PRKCZ | 5260 | 2200 | 0.352 | 0.0958 | No | ||
| 11 | CLDN15 | 16667 3572 | 2368 | 0.307 | 0.0936 | No | ||
| 12 | CLDN5 | 22826 | 2571 | 0.260 | 0.0884 | No | ||
| 13 | CSNK2A1 | 14797 | 2794 | 0.214 | 0.0812 | No | ||
| 14 | MYH7 | 21828 4709 | 2923 | 0.196 | 0.0786 | No | ||
| 15 | JAM2 | 7428 22718 12519 | 2963 | 0.192 | 0.0808 | No | ||
| 16 | INADL | 2517 2403 165 2347 166 | 3076 | 0.176 | 0.0786 | No | ||
| 17 | MYL9 | 14767 2659 | 3098 | 0.174 | 0.0814 | No | ||
| 18 | CLDN23 | 18893 | 3255 | 0.156 | 0.0764 | No | ||
| 19 | EXOC3 | 9986 | 3352 | 0.145 | 0.0744 | No | ||
| 20 | CASK | 24184 2618 2604 | 3500 | 0.131 | 0.0694 | No | ||
| 21 | VAPA | 22908 | 3566 | 0.124 | 0.0686 | No | ||
| 22 | MYH9 | 2252 2244 | 3601 | 0.122 | 0.0695 | No | ||
| 23 | PTEN | 5305 | 3651 | 0.119 | 0.0695 | No | ||
| 24 | CLDN10 | 21935 2785 2601 2720 7188 | 3834 | 0.106 | 0.0620 | No | ||
| 25 | MYL7 | 9437 | 3877 | 0.104 | 0.0621 | No | ||
| 26 | CLDN9 | 24470 | 4243 | 0.084 | 0.0442 | No | ||
| 27 | PPP2R4 | 15050 2910 | 4911 | 0.061 | 0.0095 | No | ||
| 28 | MYH11 | 22656 | 5049 | 0.057 | 0.0034 | No | ||
| 29 | CRB3 | 23176 | 5306 | 0.051 | -0.0093 | No | ||
| 30 | JAM3 | 8197 | 5839 | 0.040 | -0.0371 | No | ||
| 31 | CLDN7 | 20817 | 5943 | 0.038 | -0.0418 | No | ||
| 32 | CLDN11 | 15616 | 6223 | 0.034 | -0.0562 | No | ||
| 33 | PRKCE | 9575 | 6468 | 0.029 | -0.0687 | No | ||
| 34 | ACTG1 | 8535 | 6846 | 0.024 | -0.0885 | No | ||
| 35 | MRAS | 9409 | 7050 | 0.022 | -0.0990 | No | ||
| 36 | AKT1 | 8568 | 7183 | 0.020 | -0.1057 | No | ||
| 37 | CLDN8 | 22549 1747 | 8013 | 0.010 | -0.1502 | No | ||
| 38 | CLDN2 | 4527 | 8176 | 0.008 | -0.1588 | No | ||
| 39 | MYH3 | 20839 | 8512 | 0.005 | -0.1768 | No | ||
| 40 | CTTN | 8817 4575 1035 8818 | 8823 | 0.001 | -0.1935 | No | ||
| 41 | YES1 | 5930 | 9040 | -0.001 | -0.2051 | No | ||
| 42 | PRKCQ | 2873 2831 | 9370 | -0.004 | -0.2228 | No | ||
| 43 | MAGI1 | 19828 4816 1179 | 9454 | -0.005 | -0.2272 | No | ||
| 44 | RHOA | 8624 4409 4410 | 9584 | -0.006 | -0.2340 | No | ||
| 45 | PRKCI | 9576 | 9792 | -0.008 | -0.2450 | No | ||
| 46 | AKT2 | 4365 4366 | 9915 | -0.009 | -0.2514 | No | ||
| 47 | MYH4 | 20837 | 10179 | -0.012 | -0.2653 | No | ||
| 48 | GNAI1 | 9024 | 10182 | -0.012 | -0.2652 | No | ||
| 49 | RAB13 | 15535 | 10212 | -0.012 | -0.2665 | No | ||
| 50 | PPP2R2A | 3222 21774 | 10353 | -0.013 | -0.2737 | No | ||
| 51 | PARD6B | 12209 | 10710 | -0.018 | -0.2926 | No | ||
| 52 | PARD3 | 8212 13553 | 10948 | -0.020 | -0.3049 | No | ||
| 53 | CSNK2B | 23008 | 11034 | -0.021 | -0.3090 | No | ||
| 54 | CGN | 12896 12897 7701 | 11205 | -0.023 | -0.3177 | No | ||
| 55 | MYH2 | 20838 | 11231 | -0.024 | -0.3185 | No | ||
| 56 | RAB3B | 16141 | 11633 | -0.029 | -0.3395 | No | ||
| 57 | MYH6 | 9436 | 11876 | -0.032 | -0.3519 | No | ||
| 58 | CLDN19 | 16105 472 | 12028 | -0.034 | -0.3593 | No | ||
| 59 | MPDZ | 2497 2366 9406 | 12086 | -0.035 | -0.3616 | No | ||
| 60 | GNAI3 | 15198 | 12279 | -0.038 | -0.3711 | No | ||
| 61 | MPP5 | 21230 7078 | 12431 | -0.041 | -0.3783 | No | ||
| 62 | NRAS | 5191 | 12493 | -0.042 | -0.3807 | No | ||
| 63 | CLDN14 | 12081 | 12600 | -0.043 | -0.3855 | No | ||
| 64 | SRC | 5507 | 12732 | -0.046 | -0.3915 | No | ||
| 65 | CLDN17 | 10757 | 12817 | -0.047 | -0.3950 | No | ||
| 66 | MYH14 | 17845 | 13003 | -0.051 | -0.4039 | No | ||
| 67 | CLDN18 | 19028 | 13162 | -0.054 | -0.4112 | No | ||
| 68 | CLDN16 | 22802 | 13330 | -0.059 | -0.4189 | No | ||
| 69 | CDC42 | 4503 8722 4504 2465 | 13369 | -0.060 | -0.4197 | No | ||
| 70 | TJP1 | 17803 | 13390 | -0.060 | -0.4194 | No | ||
| 71 | PPM1J | 15466 12971 | 13763 | -0.072 | -0.4379 | No | ||
| 72 | OCLN | 21368 | 14015 | -0.082 | -0.4496 | No | ||
| 73 | TJP2 | 5761 3699 | 14043 | -0.083 | -0.4492 | No | ||
| 74 | CLDN3 | 16680 | 14046 | -0.083 | -0.4475 | No | ||
| 75 | ACTN3 | 23967 8540 | 14262 | -0.092 | -0.4571 | No | ||
| 76 | F11R | 9199 | 14381 | -0.097 | -0.4613 | No | ||
| 77 | PPP2CB | 18636 | 15083 | -0.143 | -0.4960 | No | ||
| 78 | ACTN2 | 21545 | 15268 | -0.160 | -0.5024 | No | ||
| 79 | PPP2CA | 20890 | 16118 | -0.299 | -0.5416 | No | ||
| 80 | PPP2R1A | 11951 | 16124 | -0.301 | -0.5352 | No | ||
| 81 | PARD6G | 23503 | 16409 | -0.375 | -0.5422 | Yes | ||
| 82 | PARD6A | 12142 | 16545 | -0.411 | -0.5404 | Yes | ||
| 83 | PRKCA | 20174 | 16682 | -0.460 | -0.5375 | Yes | ||
| 84 | MYH10 | 8077 | 16724 | -0.474 | -0.5292 | Yes | ||
| 85 | ASH1L | 5311 15546 15547 | 16772 | -0.497 | -0.5207 | Yes | ||
| 86 | CLDN4 | 8747 | 17169 | -0.663 | -0.5274 | Yes | ||
| 87 | KRAS | 9247 | 17394 | -0.797 | -0.5218 | Yes | ||
| 88 | HRAS | 4868 | 17672 | -0.978 | -0.5151 | Yes | ||
| 89 | HCLS1 | 22770 | 17831 | -1.068 | -0.5000 | Yes | ||
| 90 | CDK4 | 3424 19859 | 17842 | -1.076 | -0.4766 | Yes | ||
| 91 | CSDA | 1163 16966 1134 | 18129 | -1.394 | -0.4612 | Yes | ||
| 92 | PRKCH | 21246 | 18213 | -1.553 | -0.4312 | Yes | ||
| 93 | ACTB | 8534 337 337 338 | 18221 | -1.558 | -0.3970 | Yes | ||
| 94 | CSNK2A2 | 4567 8808 | 18260 | -1.597 | -0.3637 | Yes | ||
| 95 | RRAS2 | 12431 | 18490 | -2.316 | -0.3247 | Yes | ||
| 96 | EXOC4 | 17493 | 18557 | -2.919 | -0.2635 | Yes | ||
| 97 | MYLC2PL | 16670 | 18606 | -5.102 | -0.1530 | Yes | ||
| 98 | AMOTL1 | 19242 | 18612 | -6.921 | 0.0002 | Yes |