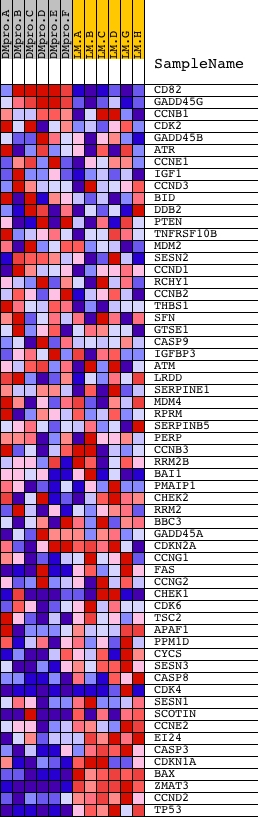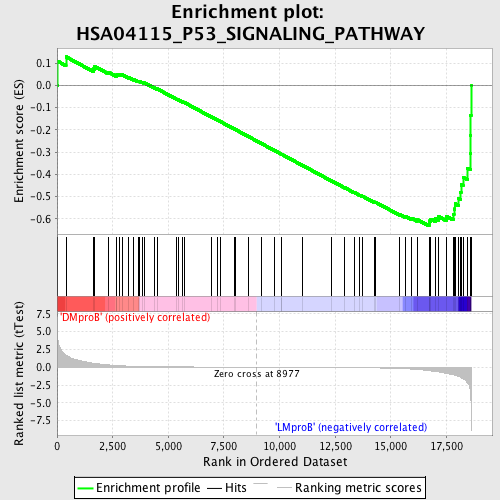
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | HSA04115_P53_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.63136894 |
| Normalized Enrichment Score (NES) | -1.5428337 |
| Nominal p-value | 0.0020491802 |
| FDR q-value | 0.52564365 |
| FWER p-Value | 0.936 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CD82 | 8720 | 17 | 4.567 | 0.1085 | No | ||
| 2 | GADD45G | 21637 | 401 | 1.662 | 0.1276 | No | ||
| 3 | CCNB1 | 11201 21362 | 1651 | 0.559 | 0.0737 | No | ||
| 4 | CDK2 | 3438 3373 19592 3322 | 1686 | 0.541 | 0.0848 | No | ||
| 5 | GADD45B | 19935 | 2318 | 0.321 | 0.0585 | No | ||
| 6 | ATR | 19349 | 2673 | 0.238 | 0.0451 | No | ||
| 7 | CCNE1 | 17857 | 2685 | 0.235 | 0.0502 | No | ||
| 8 | IGF1 | 3352 9156 3409 | 2811 | 0.211 | 0.0485 | No | ||
| 9 | CCND3 | 4489 4490 | 2917 | 0.197 | 0.0475 | No | ||
| 10 | BID | 17017 435 | 3229 | 0.158 | 0.0346 | No | ||
| 11 | DDB2 | 2721 2878 14525 2661 | 3439 | 0.137 | 0.0266 | No | ||
| 12 | PTEN | 5305 | 3651 | 0.119 | 0.0181 | No | ||
| 13 | TNFRSF10B | 21971 10205 5781 10204 | 3714 | 0.114 | 0.0174 | No | ||
| 14 | MDM2 | 19620 3327 | 3820 | 0.107 | 0.0144 | No | ||
| 15 | SESN2 | 15733 | 3933 | 0.100 | 0.0107 | No | ||
| 16 | CCND1 | 4487 4488 8707 17535 | 4372 | 0.079 | -0.0110 | No | ||
| 17 | RCHY1 | 12646 | 4499 | 0.074 | -0.0160 | No | ||
| 18 | CCNB2 | 19067 | 5368 | 0.050 | -0.0616 | No | ||
| 19 | THBS1 | 5748 14910 | 5461 | 0.047 | -0.0654 | No | ||
| 20 | SFN | 7060 12062 | 5653 | 0.044 | -0.0747 | No | ||
| 21 | GTSE1 | 11385 2170 | 5739 | 0.042 | -0.0783 | No | ||
| 22 | CASP9 | 16001 2410 2458 | 6938 | 0.023 | -0.1423 | No | ||
| 23 | IGFBP3 | 4898 | 7198 | 0.020 | -0.1558 | No | ||
| 24 | ATM | 2976 19115 | 7356 | 0.018 | -0.1638 | No | ||
| 25 | LRDD | 17555 | 7957 | 0.011 | -0.1959 | No | ||
| 26 | SERPINE1 | 16333 | 8035 | 0.010 | -0.1998 | No | ||
| 27 | MDM4 | 5086 5085 | 8620 | 0.004 | -0.2312 | No | ||
| 28 | RPRM | 14588 | 9191 | -0.002 | -0.2618 | No | ||
| 29 | SERPINB5 | 9861 | 9780 | -0.008 | -0.2933 | No | ||
| 30 | PERP | 2259 22121 20085 | 10068 | -0.011 | -0.3085 | No | ||
| 31 | CCNB3 | 24205 | 10095 | -0.011 | -0.3097 | No | ||
| 32 | RRM2B | 6902 6903 11801 | 11033 | -0.021 | -0.3597 | No | ||
| 33 | BAI1 | 22459 | 12338 | -0.039 | -0.4290 | No | ||
| 34 | PMAIP1 | 23530 12216 7196 | 12936 | -0.049 | -0.4600 | No | ||
| 35 | CHEK2 | 16751 3587 | 13353 | -0.059 | -0.4810 | No | ||
| 36 | RRM2 | 5401 5400 | 13608 | -0.067 | -0.4931 | No | ||
| 37 | BBC3 | 18382 2044 | 13733 | -0.071 | -0.4980 | No | ||
| 38 | GADD45A | 17129 | 14250 | -0.091 | -0.5237 | No | ||
| 39 | CDKN2A | 2491 15841 | 14297 | -0.093 | -0.5239 | No | ||
| 40 | CCNG1 | 4492 1236 1390 | 15406 | -0.176 | -0.5794 | No | ||
| 41 | FAS | 23881 3750 | 15661 | -0.211 | -0.5880 | No | ||
| 42 | CCNG2 | 16790 | 15942 | -0.259 | -0.5969 | No | ||
| 43 | CHEK1 | 19181 3085 | 16177 | -0.313 | -0.6021 | No | ||
| 44 | CDK6 | 16600 | 16722 | -0.473 | -0.6200 | Yes | ||
| 45 | TSC2 | 23090 | 16727 | -0.475 | -0.6089 | Yes | ||
| 46 | APAF1 | 8606 | 16795 | -0.511 | -0.6002 | Yes | ||
| 47 | PPM1D | 20721 | 17001 | -0.588 | -0.5972 | Yes | ||
| 48 | CYCS | 8821 | 17152 | -0.657 | -0.5896 | Yes | ||
| 49 | SESN3 | 19561 | 17492 | -0.860 | -0.5872 | Yes | ||
| 50 | CASP8 | 8694 | 17804 | -1.050 | -0.5788 | Yes | ||
| 51 | CDK4 | 3424 19859 | 17842 | -1.076 | -0.5551 | Yes | ||
| 52 | SESN1 | 20045 | 17884 | -1.107 | -0.5308 | Yes | ||
| 53 | SCOTIN | 3136 19299 3102 | 18047 | -1.277 | -0.5089 | Yes | ||
| 54 | CCNE2 | 4491 | 18137 | -1.408 | -0.4800 | Yes | ||
| 55 | EI24 | 19179 | 18159 | -1.436 | -0.4467 | Yes | ||
| 56 | CASP3 | 8693 | 18250 | -1.583 | -0.4136 | Yes | ||
| 57 | CDKN1A | 4511 8729 | 18443 | -2.164 | -0.3722 | Yes | ||
| 58 | BAX | 17832 | 18570 | -3.065 | -0.3056 | Yes | ||
| 59 | ZMAT3 | 10305 | 18581 | -3.349 | -0.2259 | Yes | ||
| 60 | CCND2 | 16987 | 18590 | -3.849 | -0.1341 | Yes | ||
| 61 | TP53 | 20822 | 18609 | -5.657 | 0.0004 | Yes |