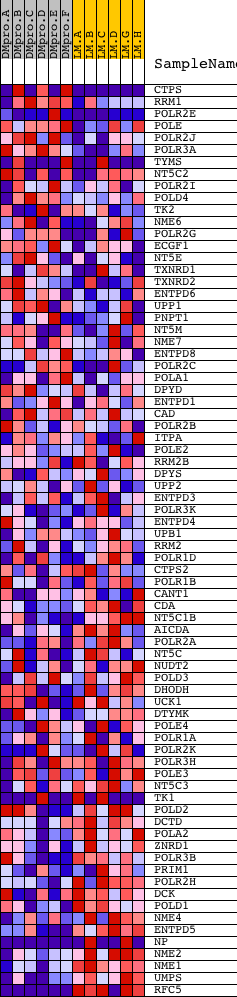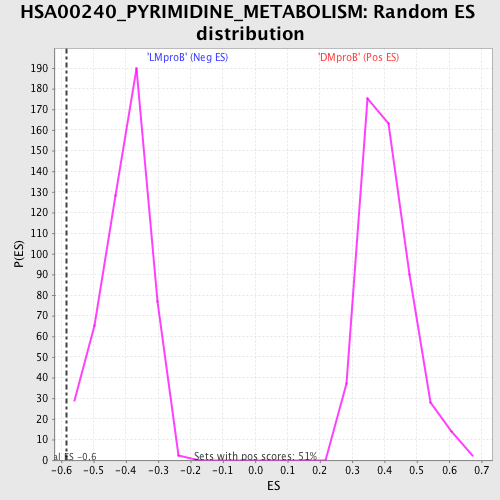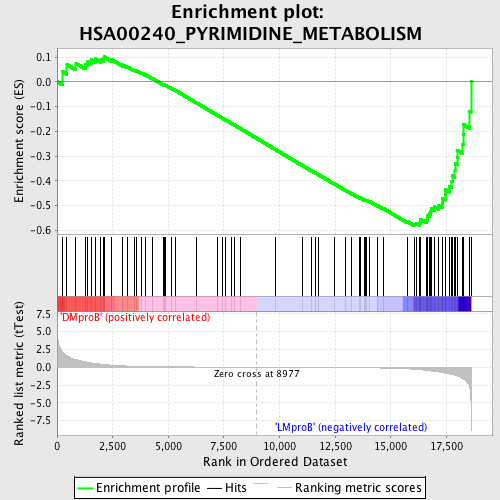
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | HSA00240_PYRIMIDINE_METABOLISM |
| Enrichment Score (ES) | -0.5828028 |
| Normalized Enrichment Score (NES) | -1.4547814 |
| Nominal p-value | 0.0061099795 |
| FDR q-value | 0.7781641 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CTPS | 2514 15772 | 226 | 2.192 | 0.0420 | No | ||
| 2 | RRM1 | 18163 | 443 | 1.598 | 0.0698 | No | ||
| 3 | POLR2E | 3325 19699 | 848 | 1.057 | 0.0742 | No | ||
| 4 | POLE | 16755 | 1260 | 0.773 | 0.0711 | No | ||
| 5 | POLR2J | 16672 | 1382 | 0.698 | 0.0818 | No | ||
| 6 | POLR3A | 21900 | 1533 | 0.612 | 0.0889 | No | ||
| 7 | TYMS | 5810 5809 3606 3598 | 1714 | 0.531 | 0.0923 | No | ||
| 8 | NT5C2 | 3768 8052 | 1969 | 0.429 | 0.0892 | No | ||
| 9 | POLR2I | 12839 | 2080 | 0.387 | 0.0928 | No | ||
| 10 | POLD4 | 12822 | 2108 | 0.380 | 0.1008 | No | ||
| 11 | TK2 | 18779 | 2453 | 0.286 | 0.0893 | No | ||
| 12 | NME6 | 3139 19297 | 2935 | 0.195 | 0.0681 | No | ||
| 13 | POLR2G | 23753 | 3165 | 0.165 | 0.0599 | No | ||
| 14 | ECGF1 | 22160 | 3461 | 0.136 | 0.0473 | No | ||
| 15 | NT5E | 19360 18702 | 3552 | 0.126 | 0.0456 | No | ||
| 16 | TXNRD1 | 19923 | 3791 | 0.109 | 0.0354 | No | ||
| 17 | TXNRD2 | 22829 1649 | 3975 | 0.098 | 0.0280 | No | ||
| 18 | ENTPD6 | 4496 2678 | 4289 | 0.082 | 0.0131 | No | ||
| 19 | UPP1 | 20947 1385 | 4796 | 0.064 | -0.0126 | No | ||
| 20 | PNPT1 | 20936 | 4812 | 0.063 | -0.0119 | No | ||
| 21 | NT5M | 8345 4175 | 4869 | 0.062 | -0.0134 | No | ||
| 22 | NME7 | 926 4101 14070 3986 3946 931 | 5139 | 0.055 | -0.0265 | No | ||
| 23 | ENTPD8 | 12996 | 5324 | 0.051 | -0.0352 | No | ||
| 24 | POLR2C | 9750 | 6273 | 0.033 | -0.0855 | No | ||
| 25 | POLA1 | 24112 | 7186 | 0.020 | -0.1342 | No | ||
| 26 | DPYD | 15437 | 7442 | 0.017 | -0.1475 | No | ||
| 27 | ENTPD1 | 4495 | 7584 | 0.015 | -0.1548 | No | ||
| 28 | CAD | 16886 | 7836 | 0.012 | -0.1680 | No | ||
| 29 | POLR2B | 16817 | 7994 | 0.011 | -0.1762 | No | ||
| 30 | ITPA | 9193 | 8226 | 0.008 | -0.1885 | No | ||
| 31 | POLE2 | 21053 | 9793 | -0.008 | -0.2727 | No | ||
| 32 | RRM2B | 6902 6903 11801 | 11033 | -0.021 | -0.3390 | No | ||
| 33 | DPYS | 22305 | 11445 | -0.026 | -0.3606 | No | ||
| 34 | UPP2 | 15009 13346 8032 | 11613 | -0.029 | -0.3689 | No | ||
| 35 | ENTPD3 | 19266 | 11767 | -0.031 | -0.3763 | No | ||
| 36 | POLR3K | 12447 7372 | 12460 | -0.041 | -0.4127 | No | ||
| 37 | ENTPD4 | 7449 | 12953 | -0.050 | -0.4380 | No | ||
| 38 | UPB1 | 19988 3389 | 13250 | -0.057 | -0.4525 | No | ||
| 39 | RRM2 | 5401 5400 | 13608 | -0.067 | -0.4701 | No | ||
| 40 | POLR1D | 3593 3658 16623 | 13620 | -0.067 | -0.4691 | No | ||
| 41 | CTPS2 | 12060 2571 | 13800 | -0.074 | -0.4769 | No | ||
| 42 | POLR1B | 14857 | 13878 | -0.077 | -0.4791 | No | ||
| 43 | CANT1 | 13304 | 13913 | -0.078 | -0.4790 | No | ||
| 44 | CDA | 15702 | 14042 | -0.083 | -0.4839 | No | ||
| 45 | NT5C1B | 21315 | 14402 | -0.098 | -0.5008 | No | ||
| 46 | AICDA | 17295 1087 | 14659 | -0.112 | -0.5119 | No | ||
| 47 | POLR2A | 5394 | 15739 | -0.224 | -0.5645 | No | ||
| 48 | NT5C | 20151 | 16079 | -0.290 | -0.5756 | Yes | ||
| 49 | NUDT2 | 16239 | 16139 | -0.303 | -0.5713 | Yes | ||
| 50 | POLD3 | 17742 | 16285 | -0.340 | -0.5707 | Yes | ||
| 51 | DHODH | 7152 | 16332 | -0.354 | -0.5645 | Yes | ||
| 52 | UCK1 | 10254 5831 10253 | 16334 | -0.355 | -0.5558 | Yes | ||
| 53 | DTYMK | 5776 | 16607 | -0.430 | -0.5598 | Yes | ||
| 54 | POLE4 | 12441 | 16649 | -0.448 | -0.5510 | Yes | ||
| 55 | POLR1A | 9749 5393 | 16667 | -0.453 | -0.5407 | Yes | ||
| 56 | POLR2K | 9413 | 16723 | -0.474 | -0.5319 | Yes | ||
| 57 | POLR3H | 13460 8135 | 16779 | -0.501 | -0.5225 | Yes | ||
| 58 | POLE3 | 7200 | 16807 | -0.514 | -0.5113 | Yes | ||
| 59 | NT5C3 | 17136 | 16967 | -0.577 | -0.5056 | Yes | ||
| 60 | TK1 | 1457 10182 5762 | 17162 | -0.661 | -0.4997 | Yes | ||
| 61 | POLD2 | 20537 | 17321 | -0.752 | -0.4897 | Yes | ||
| 62 | DCTD | 18620 | 17333 | -0.760 | -0.4715 | Yes | ||
| 63 | POLA2 | 23988 | 17453 | -0.841 | -0.4571 | Yes | ||
| 64 | ZNRD1 | 1491 22987 | 17463 | -0.845 | -0.4367 | Yes | ||
| 65 | POLR3B | 12875 | 17628 | -0.943 | -0.4222 | Yes | ||
| 66 | PRIM1 | 19847 | 17717 | -1.000 | -0.4023 | Yes | ||
| 67 | POLR2H | 10888 | 17762 | -1.025 | -0.3793 | Yes | ||
| 68 | DCK | 16808 | 17883 | -1.106 | -0.3584 | Yes | ||
| 69 | POLD1 | 17847 | 17890 | -1.117 | -0.3312 | Yes | ||
| 70 | NME4 | 23065 1577 | 17983 | -1.193 | -0.3066 | Yes | ||
| 71 | ENTPD5 | 4497 | 17993 | -1.200 | -0.2775 | Yes | ||
| 72 | NP | 22027 9597 5273 5274 | 18241 | -1.580 | -0.2517 | Yes | ||
| 73 | NME2 | 9468 | 18261 | -1.599 | -0.2132 | Yes | ||
| 74 | NME1 | 9467 | 18275 | -1.635 | -0.1735 | Yes | ||
| 75 | UMPS | 22606 1760 | 18545 | -2.771 | -0.1195 | Yes | ||
| 76 | RFC5 | 13005 7791 | 18605 | -4.988 | 0.0006 | Yes |