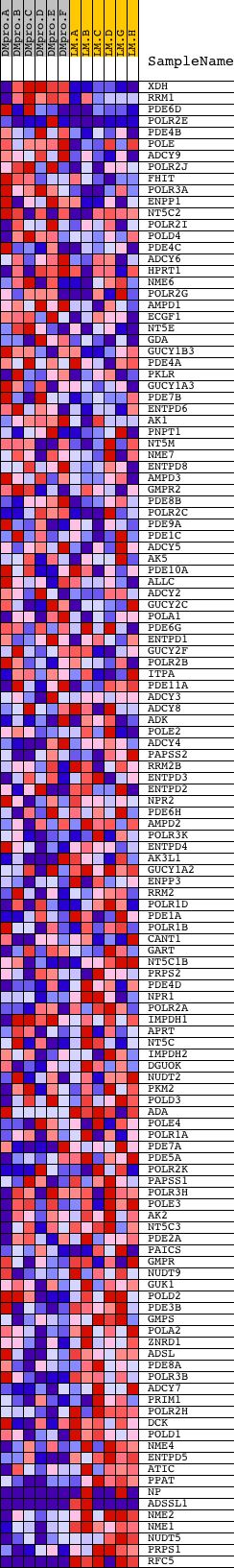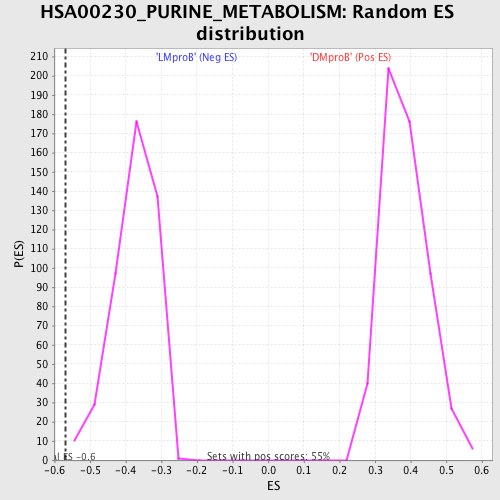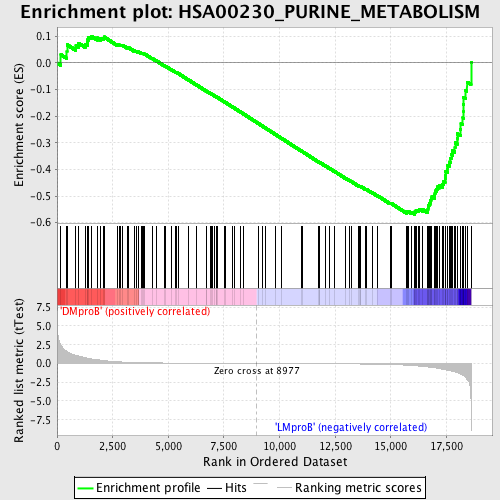
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | HSA00230_PURINE_METABOLISM |
| Enrichment Score (ES) | -0.569267 |
| Normalized Enrichment Score (NES) | -1.5154941 |
| Nominal p-value | 0.0022222223 |
| FDR q-value | 0.47260392 |
| FWER p-Value | 0.981 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | XDH | 22897 | 172 | 2.458 | 0.0311 | No | ||
| 2 | RRM1 | 18163 | 443 | 1.598 | 0.0428 | No | ||
| 3 | PDE6D | 13897 3945 | 446 | 1.588 | 0.0688 | No | ||
| 4 | POLR2E | 3325 19699 | 848 | 1.057 | 0.0644 | No | ||
| 5 | PDE4B | 9543 | 957 | 0.993 | 0.0749 | No | ||
| 6 | POLE | 16755 | 1260 | 0.773 | 0.0713 | No | ||
| 7 | ADCY9 | 22680 | 1362 | 0.708 | 0.0775 | No | ||
| 8 | POLR2J | 16672 | 1382 | 0.698 | 0.0879 | No | ||
| 9 | FHIT | 21920 | 1419 | 0.672 | 0.0970 | No | ||
| 10 | POLR3A | 21900 | 1533 | 0.612 | 0.1009 | No | ||
| 11 | ENPP1 | 19804 | 1810 | 0.495 | 0.0941 | No | ||
| 12 | NT5C2 | 3768 8052 | 1969 | 0.429 | 0.0927 | No | ||
| 13 | POLR2I | 12839 | 2080 | 0.387 | 0.0931 | No | ||
| 14 | POLD4 | 12822 | 2108 | 0.380 | 0.0979 | No | ||
| 15 | PDE4C | 8481 | 2700 | 0.231 | 0.0697 | No | ||
| 16 | ADCY6 | 22139 2283 8551 | 2802 | 0.213 | 0.0677 | No | ||
| 17 | HPRT1 | 2655 24339 408 | 2870 | 0.203 | 0.0674 | No | ||
| 18 | NME6 | 3139 19297 | 2935 | 0.195 | 0.0672 | No | ||
| 19 | POLR2G | 23753 | 3165 | 0.165 | 0.0575 | No | ||
| 20 | AMPD1 | 5051 | 3221 | 0.159 | 0.0571 | No | ||
| 21 | ECGF1 | 22160 | 3461 | 0.136 | 0.0465 | No | ||
| 22 | NT5E | 19360 18702 | 3552 | 0.126 | 0.0436 | No | ||
| 23 | GDA | 4764 | 3655 | 0.118 | 0.0401 | No | ||
| 24 | GUCY1B3 | 15311 | 3659 | 0.118 | 0.0419 | No | ||
| 25 | PDE4A | 3037 19542 3083 3023 | 3793 | 0.109 | 0.0365 | No | ||
| 26 | PKLR | 1850 15545 | 3825 | 0.107 | 0.0365 | No | ||
| 27 | GUCY1A3 | 7216 12245 15310 | 3878 | 0.104 | 0.0354 | No | ||
| 28 | PDE7B | 19807 | 3945 | 0.099 | 0.0335 | No | ||
| 29 | ENTPD6 | 4496 2678 | 4289 | 0.082 | 0.0163 | No | ||
| 30 | AK1 | 4363 | 4453 | 0.075 | 0.0087 | No | ||
| 31 | PNPT1 | 20936 | 4812 | 0.063 | -0.0096 | No | ||
| 32 | NT5M | 8345 4175 | 4869 | 0.062 | -0.0116 | No | ||
| 33 | NME7 | 926 4101 14070 3986 3946 931 | 5139 | 0.055 | -0.0253 | No | ||
| 34 | ENTPD8 | 12996 | 5324 | 0.051 | -0.0344 | No | ||
| 35 | AMPD3 | 4383 2110 | 5358 | 0.050 | -0.0354 | No | ||
| 36 | GMPR2 | 2711 | 5453 | 0.047 | -0.0397 | No | ||
| 37 | PDE8B | 5756 | 5919 | 0.039 | -0.0642 | No | ||
| 38 | POLR2C | 9750 | 6273 | 0.033 | -0.0828 | No | ||
| 39 | PDE9A | 23301 | 6696 | 0.026 | -0.1052 | No | ||
| 40 | PDE1C | 5235 | 6711 | 0.026 | -0.1055 | No | ||
| 41 | ADCY5 | 10312 | 6874 | 0.024 | -0.1139 | No | ||
| 42 | AK5 | 6042 | 6941 | 0.023 | -0.1171 | No | ||
| 43 | PDE10A | 23387 | 6965 | 0.023 | -0.1179 | No | ||
| 44 | ALLC | 21093 2130 | 7077 | 0.021 | -0.1236 | No | ||
| 45 | ADCY2 | 21412 | 7144 | 0.020 | -0.1268 | No | ||
| 46 | GUCY2C | 1140 16954 | 7164 | 0.020 | -0.1275 | No | ||
| 47 | POLA1 | 24112 | 7186 | 0.020 | -0.1283 | No | ||
| 48 | PDE6G | 20119 | 7506 | 0.016 | -0.1453 | No | ||
| 49 | ENTPD1 | 4495 | 7584 | 0.015 | -0.1492 | No | ||
| 50 | GUCY2F | 24045 | 7880 | 0.012 | -0.1650 | No | ||
| 51 | POLR2B | 16817 | 7994 | 0.011 | -0.1709 | No | ||
| 52 | ITPA | 9193 | 8226 | 0.008 | -0.1833 | No | ||
| 53 | PDE11A | 10805 | 8385 | 0.006 | -0.1917 | No | ||
| 54 | ADCY3 | 21329 894 | 9063 | -0.001 | -0.2283 | No | ||
| 55 | ADCY8 | 22281 | 9222 | -0.003 | -0.2368 | No | ||
| 56 | ADK | 3302 3454 8555 | 9381 | -0.004 | -0.2453 | No | ||
| 57 | POLE2 | 21053 | 9793 | -0.008 | -0.2674 | No | ||
| 58 | ADCY4 | 2980 21818 | 10065 | -0.011 | -0.2819 | No | ||
| 59 | PAPSS2 | 6258 | 10972 | -0.020 | -0.3306 | No | ||
| 60 | RRM2B | 6902 6903 11801 | 11033 | -0.021 | -0.3335 | No | ||
| 61 | ENTPD3 | 19266 | 11767 | -0.031 | -0.3726 | No | ||
| 62 | ENTPD2 | 8713 | 11794 | -0.031 | -0.3735 | No | ||
| 63 | NPR2 | 16230 | 11798 | -0.031 | -0.3731 | No | ||
| 64 | PDE6H | 17257 | 12060 | -0.035 | -0.3867 | No | ||
| 65 | AMPD2 | 1931 15197 | 12264 | -0.038 | -0.3970 | No | ||
| 66 | POLR3K | 12447 7372 | 12460 | -0.041 | -0.4069 | No | ||
| 67 | ENTPD4 | 7449 | 12953 | -0.050 | -0.4327 | No | ||
| 68 | AK3L1 | 8564 | 13139 | -0.054 | -0.4418 | No | ||
| 69 | GUCY1A2 | 19580 | 13226 | -0.056 | -0.4456 | No | ||
| 70 | ENPP3 | 19803 | 13530 | -0.065 | -0.4609 | No | ||
| 71 | RRM2 | 5401 5400 | 13608 | -0.067 | -0.4640 | No | ||
| 72 | POLR1D | 3593 3658 16623 | 13620 | -0.067 | -0.4634 | No | ||
| 73 | PDE1A | 9541 5234 | 13638 | -0.068 | -0.4633 | No | ||
| 74 | POLR1B | 14857 | 13878 | -0.077 | -0.4749 | No | ||
| 75 | CANT1 | 13304 | 13913 | -0.078 | -0.4755 | No | ||
| 76 | GART | 22543 1754 | 14175 | -0.088 | -0.4881 | No | ||
| 77 | NT5C1B | 21315 | 14402 | -0.098 | -0.4987 | No | ||
| 78 | PRPS2 | 24003 | 14976 | -0.134 | -0.5275 | No | ||
| 79 | PDE4D | 10722 6235 | 15012 | -0.136 | -0.5272 | No | ||
| 80 | NPR1 | 9480 | 15719 | -0.220 | -0.5618 | No | ||
| 81 | POLR2A | 5394 | 15739 | -0.224 | -0.5591 | No | ||
| 82 | IMPDH1 | 17197 1131 | 15783 | -0.231 | -0.5576 | No | ||
| 83 | APRT | 8620 | 15948 | -0.260 | -0.5622 | No | ||
| 84 | NT5C | 20151 | 16079 | -0.290 | -0.5645 | Yes | ||
| 85 | IMPDH2 | 10730 | 16083 | -0.291 | -0.5599 | Yes | ||
| 86 | DGUOK | 17099 1041 | 16105 | -0.297 | -0.5561 | Yes | ||
| 87 | NUDT2 | 16239 | 16139 | -0.303 | -0.5529 | Yes | ||
| 88 | PKM2 | 3642 9573 | 16260 | -0.334 | -0.5539 | Yes | ||
| 89 | POLD3 | 17742 | 16285 | -0.340 | -0.5497 | Yes | ||
| 90 | ADA | 2703 14361 | 16439 | -0.384 | -0.5516 | Yes | ||
| 91 | POLE4 | 12441 | 16649 | -0.448 | -0.5556 | Yes | ||
| 92 | POLR1A | 9749 5393 | 16667 | -0.453 | -0.5491 | Yes | ||
| 93 | PDE7A | 9544 1788 | 16673 | -0.458 | -0.5418 | Yes | ||
| 94 | PDE5A | 15431 | 16683 | -0.460 | -0.5347 | Yes | ||
| 95 | POLR2K | 9413 | 16723 | -0.474 | -0.5290 | Yes | ||
| 96 | PAPSS1 | 15419 | 16763 | -0.494 | -0.5230 | Yes | ||
| 97 | POLR3H | 13460 8135 | 16779 | -0.501 | -0.5156 | Yes | ||
| 98 | POLE3 | 7200 | 16807 | -0.514 | -0.5086 | Yes | ||
| 99 | AK2 | 8563 2479 16073 | 16848 | -0.535 | -0.5020 | Yes | ||
| 100 | NT5C3 | 17136 | 16967 | -0.577 | -0.4989 | Yes | ||
| 101 | PDE2A | 18170 | 16983 | -0.583 | -0.4901 | Yes | ||
| 102 | PAICS | 16820 | 16990 | -0.585 | -0.4809 | Yes | ||
| 103 | GMPR | 21651 | 17049 | -0.609 | -0.4740 | Yes | ||
| 104 | NUDT9 | 7897 | 17087 | -0.627 | -0.4657 | Yes | ||
| 105 | GUK1 | 20432 | 17167 | -0.661 | -0.4591 | Yes | ||
| 106 | POLD2 | 20537 | 17321 | -0.752 | -0.4550 | Yes | ||
| 107 | PDE3B | 5236 | 17380 | -0.788 | -0.4452 | Yes | ||
| 108 | GMPS | 15578 | 17439 | -0.831 | -0.4346 | Yes | ||
| 109 | POLA2 | 23988 | 17453 | -0.841 | -0.4215 | Yes | ||
| 110 | ZNRD1 | 1491 22987 | 17463 | -0.845 | -0.4081 | Yes | ||
| 111 | ADSL | 4358 | 17553 | -0.892 | -0.3983 | Yes | ||
| 112 | PDE8A | 18202 | 17560 | -0.895 | -0.3839 | Yes | ||
| 113 | POLR3B | 12875 | 17628 | -0.943 | -0.3720 | Yes | ||
| 114 | ADCY7 | 8552 448 | 17689 | -0.991 | -0.3590 | Yes | ||
| 115 | PRIM1 | 19847 | 17717 | -1.000 | -0.3440 | Yes | ||
| 116 | POLR2H | 10888 | 17762 | -1.025 | -0.3295 | Yes | ||
| 117 | DCK | 16808 | 17883 | -1.106 | -0.3178 | Yes | ||
| 118 | POLD1 | 17847 | 17890 | -1.117 | -0.2998 | Yes | ||
| 119 | NME4 | 23065 1577 | 17983 | -1.193 | -0.2852 | Yes | ||
| 120 | ENTPD5 | 4497 | 17993 | -1.200 | -0.2659 | Yes | ||
| 121 | ATIC | 14231 3968 | 18136 | -1.404 | -0.2505 | Yes | ||
| 122 | PPAT | 6081 | 18153 | -1.431 | -0.2279 | Yes | ||
| 123 | NP | 22027 9597 5273 5274 | 18241 | -1.580 | -0.2066 | Yes | ||
| 124 | ADSSL1 | 21136 2087 | 18254 | -1.591 | -0.1811 | Yes | ||
| 125 | NME2 | 9468 | 18261 | -1.599 | -0.1552 | Yes | ||
| 126 | NME1 | 9467 | 18275 | -1.635 | -0.1290 | Yes | ||
| 127 | NUDT5 | 15121 | 18369 | -1.880 | -0.1031 | Yes | ||
| 128 | PRPS1 | 24233 | 18424 | -2.091 | -0.0717 | Yes | ||
| 129 | RFC5 | 13005 7791 | 18605 | -4.988 | 0.0006 | Yes |