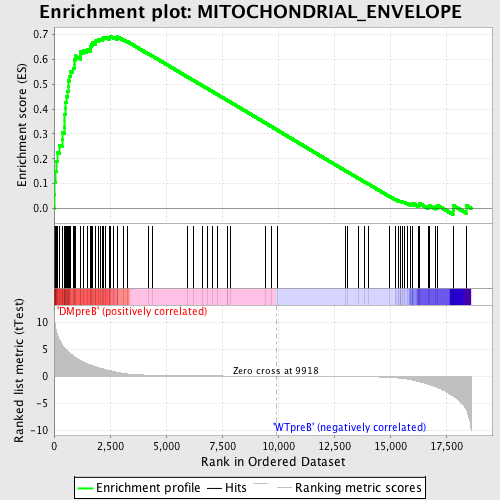
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | DMpreB |
| GeneSet | MITOCHONDRIAL_ENVELOPE |
| Enrichment Score (ES) | 0.6940726 |
| Normalized Enrichment Score (NES) | 1.6279621 |
| Nominal p-value | 0.0017152659 |
| FDR q-value | 0.021412933 |
| FWER p-Value | 0.349 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TOMM22 | 5863 | 5 | 11.381 | 0.0570 | Yes | ||
| 2 | TIMM13 | 14974 | 36 | 9.986 | 0.1057 | Yes | ||
| 3 | SLC25A11 | 20366 1368 | 77 | 8.836 | 0.1480 | Yes | ||
| 4 | ATP5G1 | 8636 | 95 | 8.444 | 0.1896 | Yes | ||
| 5 | TIMM17A | 13822 | 136 | 7.814 | 0.2268 | Yes | ||
| 6 | TIMM50 | 17911 | 245 | 6.615 | 0.2543 | Yes | ||
| 7 | BCS1L | 14223 | 355 | 5.882 | 0.2780 | Yes | ||
| 8 | PHB2 | 17286 | 367 | 5.744 | 0.3064 | Yes | ||
| 9 | TOMM34 | 14359 | 458 | 5.270 | 0.3280 | Yes | ||
| 10 | ACN9 | 12920 | 470 | 5.235 | 0.3538 | Yes | ||
| 11 | VDAC1 | 10282 | 471 | 5.220 | 0.3801 | Yes | ||
| 12 | SLC25A15 | 356 3819 | 513 | 4.994 | 0.4030 | Yes | ||
| 13 | ABCB6 | 13162 | 518 | 4.972 | 0.4278 | Yes | ||
| 14 | TIMM8B | 11391 | 549 | 4.836 | 0.4506 | Yes | ||
| 15 | ATP5G2 | 12610 | 588 | 4.682 | 0.4721 | Yes | ||
| 16 | NDUFA9 | 12288 | 639 | 4.471 | 0.4919 | Yes | ||
| 17 | BAX | 17832 | 644 | 4.463 | 0.5142 | Yes | ||
| 18 | TIMM10 | 11390 2158 | 690 | 4.300 | 0.5334 | Yes | ||
| 19 | CYCS | 8821 | 744 | 4.120 | 0.5513 | Yes | ||
| 20 | UQCRB | 12545 | 843 | 3.837 | 0.5653 | Yes | ||
| 21 | ATP5G3 | 14558 | 907 | 3.617 | 0.5801 | Yes | ||
| 22 | HTRA2 | 1122 17103 | 910 | 3.606 | 0.5982 | Yes | ||
| 23 | NDUFV1 | 23953 | 938 | 3.511 | 0.6144 | Yes | ||
| 24 | NDUFS7 | 19946 | 1179 | 2.911 | 0.6161 | Yes | ||
| 25 | ATP5O | 22539 | 1183 | 2.898 | 0.6305 | Yes | ||
| 26 | ATP5A1 | 23505 | 1292 | 2.659 | 0.6381 | Yes | ||
| 27 | SLC25A22 | 12671 | 1468 | 2.305 | 0.6403 | Yes | ||
| 28 | MPV17 | 9407 3505 3543 | 1611 | 2.112 | 0.6432 | Yes | ||
| 29 | SDHD | 19125 | 1637 | 2.073 | 0.6523 | Yes | ||
| 30 | SLC25A1 | 22645 | 1667 | 2.031 | 0.6610 | Yes | ||
| 31 | SLC25A3 | 19644 | 1730 | 1.922 | 0.6673 | Yes | ||
| 32 | SURF1 | 14645 | 1843 | 1.755 | 0.6701 | Yes | ||
| 33 | NDUFS3 | 7553 12683 | 1866 | 1.729 | 0.6776 | Yes | ||
| 34 | VDAC3 | 10284 | 1980 | 1.576 | 0.6795 | Yes | ||
| 35 | VDAC2 | 22081 | 2068 | 1.469 | 0.6822 | Yes | ||
| 36 | NDUFA2 | 9450 | 2156 | 1.374 | 0.6844 | Yes | ||
| 37 | COX15 | 5931 | 2197 | 1.316 | 0.6888 | Yes | ||
| 38 | RHOT1 | 12227 | 2298 | 1.178 | 0.6894 | Yes | ||
| 39 | NDUFAB1 | 7667 | 2456 | 1.022 | 0.6861 | Yes | ||
| 40 | ATP5J | 880 22555 | 2469 | 1.011 | 0.6905 | Yes | ||
| 41 | ATP5E | 14321 | 2497 | 0.999 | 0.6941 | Yes | ||
| 42 | NDUFA1 | 24170 | 2662 | 0.846 | 0.6895 | No | ||
| 43 | NDUFS4 | 9452 5157 3173 | 2810 | 0.721 | 0.6852 | No | ||
| 44 | NDUFS2 | 4089 13760 | 2812 | 0.719 | 0.6887 | No | ||
| 45 | TIMM9 | 19686 | 2835 | 0.698 | 0.6911 | No | ||
| 46 | ATP5D | 19949 | 3095 | 0.499 | 0.6796 | No | ||
| 47 | PMPCA | 12421 7350 | 3275 | 0.402 | 0.6720 | No | ||
| 48 | PPOX | 13761 4104 | 4204 | 0.185 | 0.6228 | No | ||
| 49 | BCL2 | 8651 3928 13864 4435 981 4062 13863 4027 | 4380 | 0.162 | 0.6142 | No | ||
| 50 | BNIP3 | 17581 | 5936 | 0.068 | 0.5306 | No | ||
| 51 | TIMM23 | 7004 5768 | 6240 | 0.059 | 0.5146 | No | ||
| 52 | MFN2 | 15678 2417 | 6621 | 0.051 | 0.4943 | No | ||
| 53 | MTX2 | 14980 | 6863 | 0.045 | 0.4815 | No | ||
| 54 | NNT | 3273 5181 9471 3238 | 7053 | 0.041 | 0.4715 | No | ||
| 55 | UCP3 | 3972 18175 | 7304 | 0.036 | 0.4582 | No | ||
| 56 | IMMT | 367 8029 | 7760 | 0.028 | 0.4338 | No | ||
| 57 | NDUFS1 | 13940 | 7761 | 0.028 | 0.4340 | No | ||
| 58 | MAOB | 24182 2556 | 7858 | 0.027 | 0.4289 | No | ||
| 59 | CENTA2 | 20748 | 9414 | 0.006 | 0.3450 | No | ||
| 60 | HCCS | 9079 4842 | 9710 | 0.003 | 0.3291 | No | ||
| 61 | SDHA | 12436 | 9969 | -0.001 | 0.3152 | No | ||
| 62 | UQCRC1 | 10259 | 12988 | -0.054 | 0.1526 | No | ||
| 63 | PHB | 9558 | 13086 | -0.057 | 0.1476 | No | ||
| 64 | OPA1 | 13169 22800 7894 | 13601 | -0.078 | 0.1203 | No | ||
| 65 | GATM | 14451 | 13842 | -0.091 | 0.1078 | No | ||
| 66 | HSD3B1 | 15227 | 14029 | -0.104 | 0.0983 | No | ||
| 67 | ATP5B | 19846 | 14988 | -0.238 | 0.0478 | No | ||
| 68 | HSD3B2 | 4873 4874 15229 4875 9126 | 15241 | -0.299 | 0.0357 | No | ||
| 69 | COX6B2 | 17983 4110 | 15358 | -0.337 | 0.0311 | No | ||
| 70 | UQCRH | 12378 | 15445 | -0.369 | 0.0283 | No | ||
| 71 | NDUFA13 | 7400 | 15469 | -0.378 | 0.0290 | No | ||
| 72 | MRPL32 | 7961 | 15546 | -0.419 | 0.0270 | No | ||
| 73 | RAB11FIP5 | 17089 | 15620 | -0.451 | 0.0253 | No | ||
| 74 | AIFM2 | 3320 3407 20006 3317 | 15765 | -0.529 | 0.0202 | No | ||
| 75 | ATP5C1 | 8635 | 15887 | -0.627 | 0.0169 | No | ||
| 76 | NDUFS8 | 23950 | 15985 | -0.705 | 0.0152 | No | ||
| 77 | ALAS2 | 24229 | 15987 | -0.706 | 0.0187 | No | ||
| 78 | RHOT2 | 1533 10073 | 16007 | -0.729 | 0.0213 | No | ||
| 79 | ABCF2 | 11338 6580 6579 | 16260 | -0.987 | 0.0127 | No | ||
| 80 | OGDH | 9502 1412 | 16302 | -1.003 | 0.0155 | No | ||
| 81 | CASP7 | 23826 | 16324 | -1.018 | 0.0195 | No | ||
| 82 | HADHB | 3514 10527 | 16691 | -1.492 | 0.0073 | No | ||
| 83 | NDUFA6 | 12473 | 16746 | -1.581 | 0.0123 | No | ||
| 84 | ABCB7 | 24085 | 17021 | -1.931 | 0.0073 | No | ||
| 85 | RAF1 | 17035 | 17131 | -2.125 | 0.0121 | No | ||
| 86 | FIS1 | 16669 3579 | 17808 | -3.640 | -0.0061 | No | ||
| 87 | TIMM17B | 24390 | 17840 | -3.710 | 0.0109 | No | ||
| 88 | MCL1 | 15502 | 18395 | -6.138 | 0.0119 | No |

