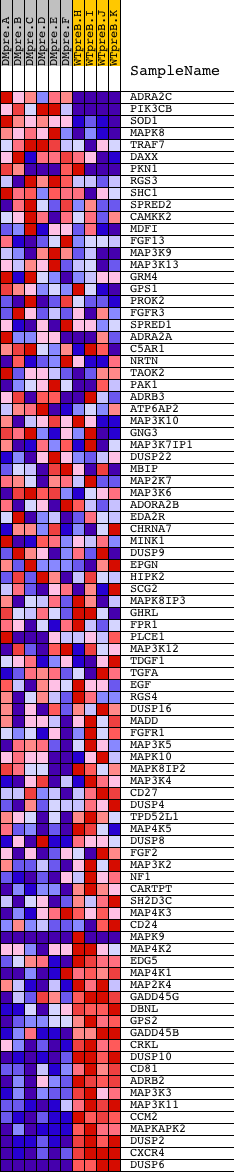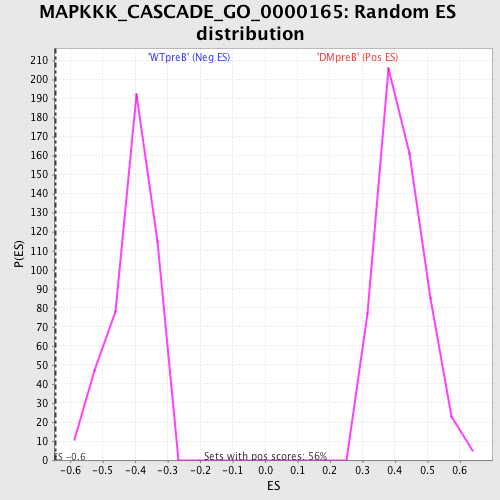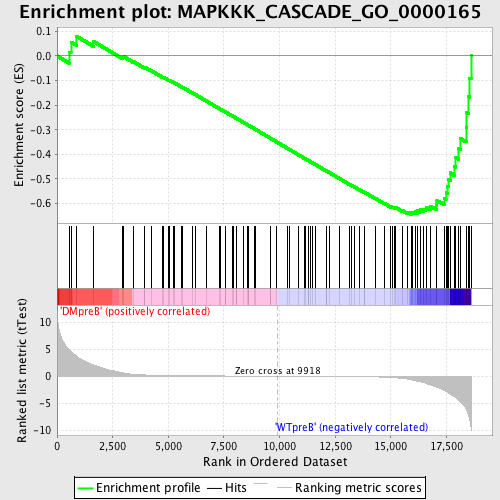
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | MAPKKK_CASCADE_GO_0000165 |
| Enrichment Score (ES) | -0.6444112 |
| Normalized Enrichment Score (NES) | -1.5798544 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.5212492 |
| FWER p-Value | 0.839 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ADRA2C | 16869 | 568 | 4.746 | 0.0155 | No | ||
| 2 | PIK3CB | 19030 | 626 | 4.520 | 0.0565 | No | ||
| 3 | SOD1 | 9846 | 863 | 3.786 | 0.0806 | No | ||
| 4 | MAPK8 | 6459 | 1644 | 2.061 | 0.0586 | No | ||
| 5 | TRAF7 | 10325 | 2928 | 0.617 | -0.0047 | No | ||
| 6 | DAXX | 23290 | 2992 | 0.562 | -0.0026 | No | ||
| 7 | PKN1 | 11567 6799 | 3449 | 0.341 | -0.0239 | No | ||
| 8 | RGS3 | 2434 2474 6937 11935 2374 2332 2445 2390 | 3930 | 0.226 | -0.0476 | No | ||
| 9 | SHC1 | 9813 9812 5430 | 3936 | 0.225 | -0.0457 | No | ||
| 10 | SPRED2 | 8539 4332 | 4238 | 0.180 | -0.0602 | No | ||
| 11 | CAMKK2 | 9870 | 4746 | 0.129 | -0.0863 | No | ||
| 12 | MDFI | 22950 | 4775 | 0.127 | -0.0866 | No | ||
| 13 | FGF13 | 4720 8963 2627 | 5013 | 0.111 | -0.0983 | No | ||
| 14 | MAP3K9 | 6872 | 5030 | 0.111 | -0.0981 | No | ||
| 15 | MAP3K13 | 22814 | 5223 | 0.100 | -0.1075 | No | ||
| 16 | GRM4 | 332 | 5282 | 0.097 | -0.1097 | No | ||
| 17 | GPS1 | 1392 9943 5550 9944 | 5598 | 0.082 | -0.1259 | No | ||
| 18 | PROK2 | 17057 | 5630 | 0.080 | -0.1268 | No | ||
| 19 | FGFR3 | 8969 3566 | 6071 | 0.064 | -0.1499 | No | ||
| 20 | SPRED1 | 4331 | 6232 | 0.060 | -0.1579 | No | ||
| 21 | ADRA2A | 4355 | 6721 | 0.048 | -0.1838 | No | ||
| 22 | C5AR1 | 17957 | 7290 | 0.037 | -0.2141 | No | ||
| 23 | NRTN | 22923 | 7344 | 0.036 | -0.2166 | No | ||
| 24 | TAOK2 | 10610 3794 | 7576 | 0.031 | -0.2288 | No | ||
| 25 | PAK1 | 9527 | 7590 | 0.031 | -0.2292 | No | ||
| 26 | ADRB3 | 18901 | 7882 | 0.027 | -0.2446 | No | ||
| 27 | ATP6AP2 | 2567 24380 2579 2656 | 7946 | 0.026 | -0.2478 | No | ||
| 28 | MAP3K10 | 3831 17916 | 8049 | 0.024 | -0.2530 | No | ||
| 29 | GNG3 | 23752 | 8071 | 0.024 | -0.2539 | No | ||
| 30 | MAP3K7IP1 | 2193 2171 22419 | 8368 | 0.020 | -0.2697 | No | ||
| 31 | DUSP22 | 466 21680 | 8571 | 0.017 | -0.2805 | No | ||
| 32 | MBIP | 21063 2072 | 8582 | 0.017 | -0.2808 | No | ||
| 33 | MAP2K7 | 6453 | 8613 | 0.016 | -0.2823 | No | ||
| 34 | MAP3K6 | 16054 | 8852 | 0.014 | -0.2950 | No | ||
| 35 | ADORA2B | 20850 | 8922 | 0.013 | -0.2986 | No | ||
| 36 | EDA2R | 6364 | 9593 | 0.004 | -0.3347 | No | ||
| 37 | CHRNA7 | 17808 | 9599 | 0.004 | -0.3350 | No | ||
| 38 | MINK1 | 20802 | 9865 | 0.001 | -0.3493 | No | ||
| 39 | DUSP9 | 24307 | 10373 | -0.005 | -0.3766 | No | ||
| 40 | EPGN | 16798 | 10442 | -0.006 | -0.3802 | No | ||
| 41 | HIPK2 | 17180 | 10830 | -0.012 | -0.4010 | No | ||
| 42 | SCG2 | 5410 14425 | 11138 | -0.016 | -0.4174 | No | ||
| 43 | MAPK8IP3 | 11402 23080 | 11152 | -0.016 | -0.4179 | No | ||
| 44 | GHRL | 1183 17040 | 11284 | -0.018 | -0.4248 | No | ||
| 45 | FPR1 | 23115 | 11300 | -0.019 | -0.4255 | No | ||
| 46 | PLCE1 | 13156 | 11374 | -0.020 | -0.4292 | No | ||
| 47 | MAP3K12 | 6454 11164 | 11465 | -0.021 | -0.4339 | No | ||
| 48 | TDGF1 | 5704 10108 | 11624 | -0.024 | -0.4422 | No | ||
| 49 | TGFA | 17381 | 12092 | -0.032 | -0.4671 | No | ||
| 50 | EGF | 15169 | 12130 | -0.033 | -0.4687 | No | ||
| 51 | RGS4 | 9724 | 12233 | -0.035 | -0.4739 | No | ||
| 52 | DUSP16 | 1004 7699 | 12674 | -0.046 | -0.4972 | No | ||
| 53 | MADD | 2926 14526 | 13141 | -0.058 | -0.5218 | No | ||
| 54 | FGFR1 | 3789 8968 | 13230 | -0.062 | -0.5259 | No | ||
| 55 | MAP3K5 | 20079 11166 | 13364 | -0.067 | -0.5324 | No | ||
| 56 | MAPK10 | 11169 | 13581 | -0.077 | -0.5434 | No | ||
| 57 | MAPK8IP2 | 77 | 13821 | -0.090 | -0.5554 | No | ||
| 58 | MAP3K4 | 23126 | 13836 | -0.091 | -0.5553 | No | ||
| 59 | CD27 | 16994 | 14310 | -0.131 | -0.5795 | No | ||
| 60 | DUSP4 | 18632 3820 | 14695 | -0.182 | -0.5985 | No | ||
| 61 | TPD52L1 | 3420 19793 | 15003 | -0.240 | -0.6127 | No | ||
| 62 | MAP4K5 | 11853 21048 2101 2142 | 15096 | -0.263 | -0.6151 | No | ||
| 63 | DUSP8 | 9493 | 15165 | -0.280 | -0.6160 | No | ||
| 64 | FGF2 | 15608 | 15208 | -0.291 | -0.6155 | No | ||
| 65 | MAP3K2 | 11165 | 15532 | -0.412 | -0.6289 | No | ||
| 66 | NF1 | 5165 | 15767 | -0.530 | -0.6364 | No | ||
| 67 | CARTPT | 21374 | 15917 | -0.652 | -0.6381 | Yes | ||
| 68 | SH2D3C | 2864 | 15995 | -0.713 | -0.6353 | Yes | ||
| 69 | MAP4K3 | 22887 | 16091 | -0.815 | -0.6325 | Yes | ||
| 70 | CD24 | 8711 | 16207 | -0.934 | -0.6296 | Yes | ||
| 71 | MAPK9 | 1233 20903 1383 | 16314 | -1.011 | -0.6255 | Yes | ||
| 72 | MAP4K2 | 6457 | 16470 | -1.178 | -0.6224 | Yes | ||
| 73 | EDG5 | 19216 | 16581 | -1.322 | -0.6154 | Yes | ||
| 74 | MAP4K1 | 18313 | 16775 | -1.624 | -0.6100 | Yes | ||
| 75 | MAP2K4 | 20405 | 17051 | -1.981 | -0.6056 | Yes | ||
| 76 | GADD45G | 21637 | 17072 | -2.018 | -0.5870 | Yes | ||
| 77 | DBNL | 20954 | 17397 | -2.642 | -0.5788 | Yes | ||
| 78 | GPS2 | 12096 | 17482 | -2.830 | -0.5558 | Yes | ||
| 79 | GADD45B | 19935 | 17545 | -2.955 | -0.5303 | Yes | ||
| 80 | CRKL | 4560 | 17588 | -3.053 | -0.5029 | Yes | ||
| 81 | DUSP10 | 4003 14016 | 17669 | -3.295 | -0.4751 | Yes | ||
| 82 | CD81 | 8719 | 17871 | -3.794 | -0.4490 | Yes | ||
| 83 | ADRB2 | 23422 | 17922 | -3.954 | -0.4132 | Yes | ||
| 84 | MAP3K3 | 20626 | 18041 | -4.417 | -0.3766 | Yes | ||
| 85 | MAP3K11 | 11163 | 18117 | -4.677 | -0.3351 | Yes | ||
| 86 | CCM2 | 20950 | 18384 | -6.050 | -0.2906 | Yes | ||
| 87 | MAPKAPK2 | 13838 | 18416 | -6.306 | -0.2309 | Yes | ||
| 88 | DUSP2 | 14865 | 18496 | -7.254 | -0.1645 | Yes | ||
| 89 | CXCR4 | 13844 | 18551 | -7.968 | -0.0898 | Yes | ||
| 90 | DUSP6 | 19891 3399 | 18608 | -9.583 | 0.0004 | Yes |