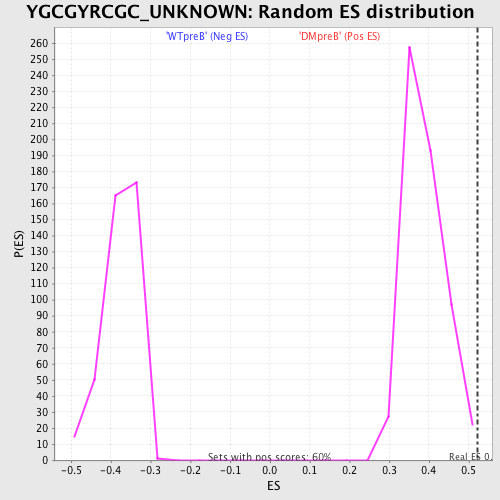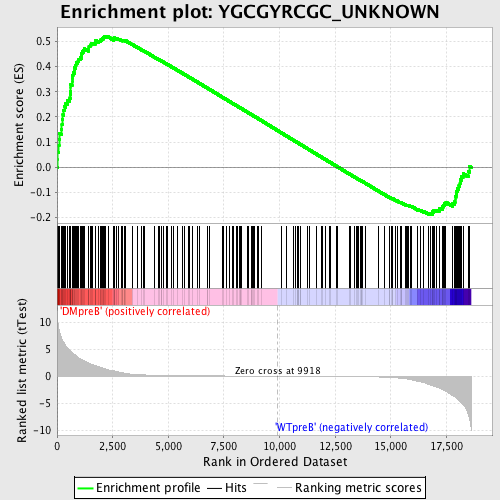
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | DMpreB |
| GeneSet | YGCGYRCGC_UNKNOWN |
| Enrichment Score (ES) | 0.5207424 |
| Normalized Enrichment Score (NES) | 1.3416398 |
| Nominal p-value | 0.0067114094 |
| FDR q-value | 0.3412311 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | IKBKAP | 2540 6045 | 22 | 10.575 | 0.0307 | Yes | ||
| 2 | TADA2L | 20316 1435 | 27 | 10.199 | 0.0612 | Yes | ||
| 3 | SLC25A11 | 20366 1368 | 77 | 8.836 | 0.0852 | Yes | ||
| 4 | AATF | 20315 | 88 | 8.545 | 0.1104 | Yes | ||
| 5 | ATP2A2 | 4421 3481 | 109 | 8.185 | 0.1340 | Yes | ||
| 6 | RPP38 | 14698 | 191 | 7.125 | 0.1511 | Yes | ||
| 7 | IMMP2L | 21283 | 217 | 6.865 | 0.1704 | Yes | ||
| 8 | PLOD3 | 16666 | 229 | 6.720 | 0.1901 | Yes | ||
| 9 | CCNE1 | 17857 | 261 | 6.545 | 0.2081 | Yes | ||
| 10 | PSMD2 | 10137 5724 | 288 | 6.286 | 0.2256 | Yes | ||
| 11 | SFRS6 | 14751 | 326 | 6.012 | 0.2418 | Yes | ||
| 12 | MTHFD2 | 17100 | 389 | 5.587 | 0.2552 | Yes | ||
| 13 | RQCD1 | 12205 | 475 | 5.201 | 0.2663 | Yes | ||
| 14 | XPO5 | 7802 | 575 | 4.724 | 0.2752 | Yes | ||
| 15 | ATP5G2 | 12610 | 588 | 4.682 | 0.2886 | Yes | ||
| 16 | RBKS | 16568 | 600 | 4.621 | 0.3020 | Yes | ||
| 17 | GEMIN4 | 6592 6591 | 612 | 4.590 | 0.3152 | Yes | ||
| 18 | RUTBC3 | 8373 | 614 | 4.586 | 0.3290 | Yes | ||
| 19 | TUBG1 | 20662 | 674 | 4.381 | 0.3390 | Yes | ||
| 20 | SLC16A6 | 4186 | 679 | 4.358 | 0.3519 | Yes | ||
| 21 | PPARGC1B | 9318 | 687 | 4.318 | 0.3645 | Yes | ||
| 22 | HS2ST1 | 15150 | 717 | 4.201 | 0.3756 | Yes | ||
| 23 | FUS | 6135 | 784 | 3.989 | 0.3841 | Yes | ||
| 24 | SKP2 | 6576 11337 2236 | 791 | 3.963 | 0.3957 | Yes | ||
| 25 | ACACA | 309 4209 | 836 | 3.856 | 0.4049 | Yes | ||
| 26 | CCS | 23966 | 851 | 3.817 | 0.4157 | Yes | ||
| 27 | PRKAR2A | 5287 | 936 | 3.521 | 0.4217 | Yes | ||
| 28 | EIF3S7 | 22226 | 975 | 3.397 | 0.4299 | Yes | ||
| 29 | APRIN | 4145 | 1045 | 3.198 | 0.4358 | Yes | ||
| 30 | TIAM1 | 22547 | 1097 | 3.076 | 0.4423 | Yes | ||
| 31 | ANP32A | 8592 4387 19415 | 1107 | 3.050 | 0.4510 | Yes | ||
| 32 | AKAP8 | 12121 | 1126 | 3.019 | 0.4591 | Yes | ||
| 33 | ARRB2 | 20806 | 1172 | 2.927 | 0.4655 | Yes | ||
| 34 | ENDOGL1 | 3115 19275 | 1238 | 2.779 | 0.4703 | Yes | ||
| 35 | FBXW9 | 18540 | 1407 | 2.403 | 0.4685 | Yes | ||
| 36 | PRKAG2 | 24 | 1431 | 2.370 | 0.4744 | Yes | ||
| 37 | ZNHIT1 | 16334 | 1432 | 2.369 | 0.4815 | Yes | ||
| 38 | RAB33A | 358 | 1484 | 2.269 | 0.4856 | Yes | ||
| 39 | GNAS | 9025 2963 2752 | 1524 | 2.229 | 0.4902 | Yes | ||
| 40 | BAT3 | 5896 | 1608 | 2.115 | 0.4921 | Yes | ||
| 41 | UROD | 15791 | 1707 | 1.954 | 0.4926 | Yes | ||
| 42 | PPM1A | 5279 9607 | 1718 | 1.933 | 0.4979 | Yes | ||
| 43 | YWHAE | 20776 | 1727 | 1.923 | 0.5033 | Yes | ||
| 44 | HNRPAB | 9104 | 1870 | 1.717 | 0.5007 | Yes | ||
| 45 | CRSP7 | 18838 | 1927 | 1.630 | 0.5026 | Yes | ||
| 46 | TFRC | 22785 | 1964 | 1.583 | 0.5054 | Yes | ||
| 47 | CFL1 | 4516 | 1984 | 1.573 | 0.5091 | Yes | ||
| 48 | RBBP4 | 5364 | 2042 | 1.502 | 0.5106 | Yes | ||
| 49 | POP7 | 16329 | 2069 | 1.468 | 0.5136 | Yes | ||
| 50 | NEK8 | 20338 | 2090 | 1.449 | 0.5169 | Yes | ||
| 51 | USP37 | 13923 | 2107 | 1.433 | 0.5203 | Yes | ||
| 52 | SEP15 | 13527 1895 15398 | 2175 | 1.346 | 0.5207 | Yes | ||
| 53 | PIM2 | 5249 9564 9565 | 2296 | 1.182 | 0.5178 | No | ||
| 54 | AAMP | 10406 | 2549 | 0.973 | 0.5070 | No | ||
| 55 | HNRPL | 18315 | 2564 | 0.961 | 0.5092 | No | ||
| 56 | VKORC1L1 | 102 7625 | 2566 | 0.960 | 0.5120 | No | ||
| 57 | RAB10 | 2090 21119 | 2585 | 0.939 | 0.5139 | No | ||
| 58 | AOF2 | 13625 | 2652 | 0.859 | 0.5129 | No | ||
| 59 | CHD4 | 8418 17281 4225 | 2762 | 0.754 | 0.5092 | No | ||
| 60 | HOOK3 | 18903 6734 | 2906 | 0.628 | 0.5034 | No | ||
| 61 | MAPT | 9420 1347 | 2946 | 0.604 | 0.5031 | No | ||
| 62 | SLC35B3 | 3225 21487 | 2959 | 0.590 | 0.5042 | No | ||
| 63 | CDK5R1 | 20745 | 3007 | 0.553 | 0.5033 | No | ||
| 64 | PAQR4 | 23105 | 3014 | 0.549 | 0.5046 | No | ||
| 65 | ACSL4 | 24043 11936 24042 2614 6940 | 3094 | 0.499 | 0.5019 | No | ||
| 66 | POLH | 22966 | 3377 | 0.361 | 0.4876 | No | ||
| 67 | UBE2S | 17979 | 3627 | 0.293 | 0.4750 | No | ||
| 68 | ADAM12 | 2207 8545 | 3812 | 0.249 | 0.4658 | No | ||
| 69 | SOX12 | 5478 | 3873 | 0.236 | 0.4632 | No | ||
| 70 | SYNCRIP | 3078 3035 3107 | 3939 | 0.225 | 0.4604 | No | ||
| 71 | COL25A1 | 8056 1837 1844 1915 13376 | 4366 | 0.164 | 0.4378 | No | ||
| 72 | MAGI1 | 19828 4816 1179 | 4552 | 0.145 | 0.4281 | No | ||
| 73 | TIMELESS | 10177 | 4598 | 0.141 | 0.4261 | No | ||
| 74 | MPP5 | 21230 7078 | 4682 | 0.134 | 0.4220 | No | ||
| 75 | DLGAP4 | 2950 10461 | 4708 | 0.132 | 0.4211 | No | ||
| 76 | ZZZ3 | 15389 8445 4253 8444 1761 | 4776 | 0.126 | 0.4178 | No | ||
| 77 | NPAS2 | 5186 | 4919 | 0.117 | 0.4105 | No | ||
| 78 | SEMA6A | 23439 9802 | 4940 | 0.116 | 0.4097 | No | ||
| 79 | TRIM33 | 8236 13579 15472 8237 | 5159 | 0.104 | 0.3982 | No | ||
| 80 | GBX2 | 13887 | 5218 | 0.100 | 0.3954 | No | ||
| 81 | ACCN2 | 22363 | 5425 | 0.089 | 0.3845 | No | ||
| 82 | RPS4X | 9756 | 5613 | 0.081 | 0.3745 | No | ||
| 83 | PARK2 | 6944 11939 1499 23385 | 5744 | 0.075 | 0.3677 | No | ||
| 84 | STARD10 | 18171 2133 | 5902 | 0.070 | 0.3594 | No | ||
| 85 | SPRY4 | 23448 | 5960 | 0.068 | 0.3565 | No | ||
| 86 | BCLAF1 | 3340 7820 | 6085 | 0.064 | 0.3500 | No | ||
| 87 | HOXA1 | 1005 17152 | 6290 | 0.058 | 0.3391 | No | ||
| 88 | EN1 | 387 14155 | 6404 | 0.055 | 0.3331 | No | ||
| 89 | SLITRK4 | 24144 | 6773 | 0.047 | 0.3133 | No | ||
| 90 | WBSCR17 | 16357 | 6775 | 0.047 | 0.3134 | No | ||
| 91 | FMR1 | 24321 | 6848 | 0.046 | 0.3096 | No | ||
| 92 | ASCL1 | 19655 | 7419 | 0.034 | 0.2788 | No | ||
| 93 | HNRPD | 8645 | 7481 | 0.033 | 0.2755 | No | ||
| 94 | STX12 | 4140 | 7622 | 0.031 | 0.2680 | No | ||
| 95 | IMMT | 367 8029 | 7760 | 0.028 | 0.2607 | No | ||
| 96 | NR2F2 | 8619 409 | 7865 | 0.027 | 0.2551 | No | ||
| 97 | KCNN1 | 8200 | 7914 | 0.026 | 0.2526 | No | ||
| 98 | LENG8 | 6114 | 8069 | 0.024 | 0.2443 | No | ||
| 99 | MNT | 20784 | 8098 | 0.023 | 0.2429 | No | ||
| 100 | CRY1 | 19662 | 8197 | 0.022 | 0.2376 | No | ||
| 101 | SORCS3 | 7323 | 8229 | 0.022 | 0.2360 | No | ||
| 102 | RHOA | 8624 4409 4410 | 8261 | 0.021 | 0.2344 | No | ||
| 103 | PHYHIPL | 12907 7711 | 8283 | 0.021 | 0.2333 | No | ||
| 104 | POU4F1 | 21727 | 8564 | 0.017 | 0.2182 | No | ||
| 105 | SLC12A5 | 14731 | 8596 | 0.016 | 0.2165 | No | ||
| 106 | CKS2 | 7264 | 8727 | 0.015 | 0.2095 | No | ||
| 107 | UBTF | 1313 10052 | 8751 | 0.015 | 0.2083 | No | ||
| 108 | MAZ | 1327 17623 | 8787 | 0.014 | 0.2064 | No | ||
| 109 | ANAPC10 | 7595 12760 7596 | 8830 | 0.014 | 0.2042 | No | ||
| 110 | WAC | 5905 5906 | 8890 | 0.013 | 0.2010 | No | ||
| 111 | TPR | 927 4255 | 8987 | 0.012 | 0.1959 | No | ||
| 112 | RGS7 | 13743 | 9046 | 0.011 | 0.1928 | No | ||
| 113 | PGM2L1 | 7712 | 9171 | 0.009 | 0.1861 | No | ||
| 114 | NEUROD1 | 14550 | 10083 | -0.002 | 0.1366 | No | ||
| 115 | GOLGA2 | 15039 2779 2923 | 10331 | -0.005 | 0.1232 | No | ||
| 116 | ZIC2 | 21927 | 10646 | -0.009 | 0.1062 | No | ||
| 117 | HOXA4 | 17148 | 10724 | -0.010 | 0.1021 | No | ||
| 118 | KCNC1 | 9204 | 10797 | -0.011 | 0.0982 | No | ||
| 119 | MMP16 | 5109 | 10818 | -0.012 | 0.0971 | No | ||
| 120 | NXPH3 | 20281 | 10829 | -0.012 | 0.0966 | No | ||
| 121 | DNAJA1 | 4878 16242 2345 | 10832 | -0.012 | 0.0966 | No | ||
| 122 | GTF2A1 | 2064 8187 8188 13512 | 10949 | -0.013 | 0.0903 | No | ||
| 123 | CYP1B1 | 4581 | 10955 | -0.013 | 0.0901 | No | ||
| 124 | RNF149 | 12570 | 11261 | -0.018 | 0.0736 | No | ||
| 125 | HNRPDL | 11947 3551 6954 | 11342 | -0.019 | 0.0693 | No | ||
| 126 | CDON | 12195 | 11675 | -0.024 | 0.0513 | No | ||
| 127 | POLR2B | 16817 | 11867 | -0.028 | 0.0411 | No | ||
| 128 | ABCE1 | 6270 | 11927 | -0.029 | 0.0379 | No | ||
| 129 | SIX1 | 9821 | 12055 | -0.031 | 0.0311 | No | ||
| 130 | INHBB | 13858 | 12235 | -0.035 | 0.0215 | No | ||
| 131 | PMP22 | 9594 1312 | 12282 | -0.036 | 0.0191 | No | ||
| 132 | NUDT3 | 23056 | 12550 | -0.043 | 0.0048 | No | ||
| 133 | NEURL | 5164 | 12583 | -0.044 | 0.0032 | No | ||
| 134 | JPH4 | 21826 | 13126 | -0.058 | -0.0261 | No | ||
| 135 | MLLT7 | 24278 | 13187 | -0.061 | -0.0291 | No | ||
| 136 | MST1 | 9089 | 13347 | -0.067 | -0.0376 | No | ||
| 137 | LRFN5 | 6213 | 13446 | -0.070 | -0.0427 | No | ||
| 138 | RPS6KA5 | 2076 21005 | 13512 | -0.073 | -0.0460 | No | ||
| 139 | GABRA4 | 16519 | 13536 | -0.075 | -0.0470 | No | ||
| 140 | KCNS2 | 4949 2232 | 13656 | -0.081 | -0.0532 | No | ||
| 141 | A2BP1 | 11212 1652 1689 | 13688 | -0.083 | -0.0547 | No | ||
| 142 | MLLT10 | 9393 5104 | 13700 | -0.083 | -0.0550 | No | ||
| 143 | PACRG | 23127 | 13713 | -0.084 | -0.0554 | No | ||
| 144 | KAZALD1 | 4206 | 13853 | -0.092 | -0.0627 | No | ||
| 145 | SHH | 16581 | 14447 | -0.146 | -0.0944 | No | ||
| 146 | MRPL45 | 20680 | 14727 | -0.188 | -0.1090 | No | ||
| 147 | FUT8 | 2163 21233 | 14949 | -0.227 | -0.1203 | No | ||
| 148 | PCTK1 | 24369 | 14960 | -0.230 | -0.1202 | No | ||
| 149 | U2AF2 | 5813 5812 5814 | 15014 | -0.242 | -0.1223 | No | ||
| 150 | POU2F1 | 5275 3989 4065 4010 | 15082 | -0.259 | -0.1252 | No | ||
| 151 | NR2C2 | 10216 | 15085 | -0.260 | -0.1245 | No | ||
| 152 | PCBP2 | 9533 | 15213 | -0.292 | -0.1305 | No | ||
| 153 | DLL1 | 23119 | 15281 | -0.313 | -0.1332 | No | ||
| 154 | HIC2 | 7185 | 15435 | -0.365 | -0.1404 | No | ||
| 155 | HNRPR | 13196 7909 2500 | 15460 | -0.375 | -0.1406 | No | ||
| 156 | ROM1 | 23751 | 15482 | -0.384 | -0.1406 | No | ||
| 157 | SPINT2 | 4128 17899 | 15643 | -0.462 | -0.1478 | No | ||
| 158 | TSGA10 | 4123 13975 5587 | 15713 | -0.501 | -0.1501 | No | ||
| 159 | RNF38 | 7862 2328 13115 | 15736 | -0.511 | -0.1497 | No | ||
| 160 | RAB6A | 18173 3905 | 15776 | -0.535 | -0.1502 | No | ||
| 161 | MTMR3 | 7908 13193 | 15872 | -0.614 | -0.1535 | No | ||
| 162 | CTDSPL | 19279 | 15922 | -0.656 | -0.1542 | No | ||
| 163 | HNRPH1 | 12222 1486 1220 7202 7201 | 16220 | -0.944 | -0.1675 | No | ||
| 164 | AP3S1 | 8604 | 16327 | -1.021 | -0.1702 | No | ||
| 165 | JMJD2C | 16188 | 16475 | -1.189 | -0.1746 | No | ||
| 166 | BCL9 | 15232 | 16684 | -1.483 | -0.1814 | No | ||
| 167 | FHOD1 | 18772 | 16792 | -1.656 | -0.1822 | No | ||
| 168 | GTF2I | 16353 5491 9863 3280 3184 | 16882 | -1.744 | -0.1818 | No | ||
| 169 | MRPL14 | 23223 | 16884 | -1.746 | -0.1766 | No | ||
| 170 | BRE | 3640 4226 16879 | 16918 | -1.795 | -0.1729 | No | ||
| 171 | PPP3CC | 21763 | 16961 | -1.850 | -0.1697 | No | ||
| 172 | CALM3 | 8682 | 17069 | -2.011 | -0.1694 | No | ||
| 173 | SLC25A12 | 14562 | 17171 | -2.189 | -0.1683 | No | ||
| 174 | GOPC | 8252 | 17176 | -2.195 | -0.1619 | No | ||
| 175 | MECP2 | 5088 | 17315 | -2.463 | -0.1619 | No | ||
| 176 | DDIT3 | 19857 | 17317 | -2.466 | -0.1546 | No | ||
| 177 | H1FX | 10835 | 17374 | -2.596 | -0.1498 | No | ||
| 178 | TMEM24 | 19150 | 17423 | -2.686 | -0.1443 | No | ||
| 179 | NAGK | 17388 1099 1121 | 17476 | -2.808 | -0.1387 | No | ||
| 180 | HIBADH | 17144 | 17782 | -3.569 | -0.1444 | No | ||
| 181 | PDE7A | 9544 1788 | 17861 | -3.771 | -0.1373 | No | ||
| 182 | DCTN4 | 12560 7474 | 17888 | -3.850 | -0.1271 | No | ||
| 183 | SRR | 6566 11328 | 17890 | -3.852 | -0.1156 | No | ||
| 184 | VRK3 | 13 | 17933 | -3.990 | -0.1058 | No | ||
| 185 | TRIB2 | 21102 | 17955 | -4.082 | -0.0946 | No | ||
| 186 | TCTA | 4167 | 18001 | -4.223 | -0.0844 | No | ||
| 187 | TIGD2 | 17429 | 18061 | -4.507 | -0.0740 | No | ||
| 188 | ULK2 | 6617 11384 | 18108 | -4.648 | -0.0625 | No | ||
| 189 | MAP3K11 | 11163 | 18117 | -4.677 | -0.0488 | No | ||
| 190 | RRAS | 18256 | 18165 | -4.858 | -0.0367 | No | ||
| 191 | TRIM8 | 23824 | 18247 | -5.292 | -0.0251 | No | ||
| 192 | KLF13 | 17807 | 18488 | -7.203 | -0.0165 | No | ||
| 193 | ZDHHC14 | 5886 | 18535 | -7.748 | 0.0044 | No |