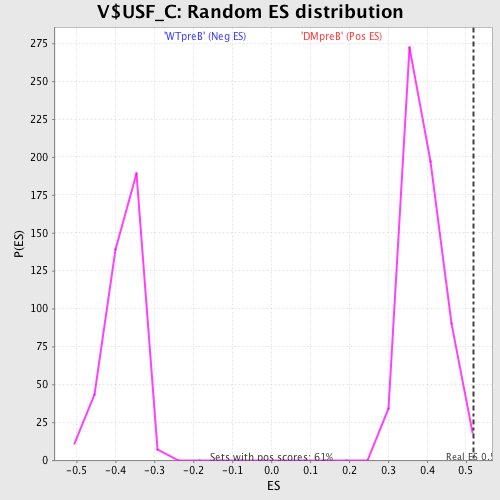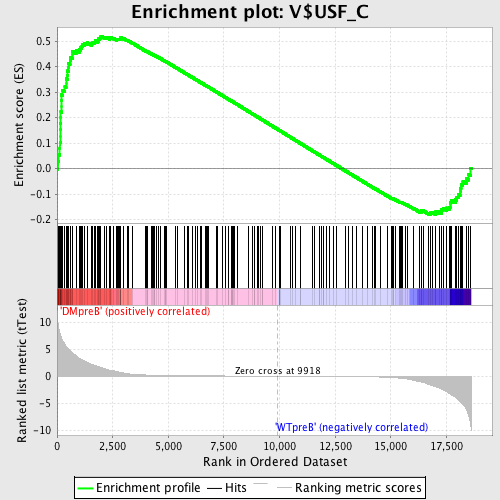
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | DMpreB |
| GeneSet | V$USF_C |
| Enrichment Score (ES) | 0.5195535 |
| Normalized Enrichment Score (NES) | 1.3305858 |
| Nominal p-value | 0.004909984 |
| FDR q-value | 0.2932772 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | FBL | 8955 | 39 | 9.764 | 0.0290 | Yes | ||
| 2 | LYAR | 16856 | 63 | 9.035 | 0.0565 | Yes | ||
| 3 | PPAT | 6081 | 124 | 7.966 | 0.0787 | Yes | ||
| 4 | IPO4 | 7983 | 129 | 7.883 | 0.1036 | Yes | ||
| 5 | EEF1E1 | 21488 | 139 | 7.768 | 0.1278 | Yes | ||
| 6 | PA2G4 | 5265 | 141 | 7.747 | 0.1525 | Yes | ||
| 7 | NUDC | 9495 | 146 | 7.691 | 0.1768 | Yes | ||
| 8 | CNOT4 | 7010 1022 | 166 | 7.395 | 0.1993 | Yes | ||
| 9 | EXOSC5 | 18334 | 171 | 7.337 | 0.2225 | Yes | ||
| 10 | EIF5A | 11345 20379 6590 | 188 | 7.145 | 0.2444 | Yes | ||
| 11 | HSPE1 | 9133 | 190 | 7.125 | 0.2670 | Yes | ||
| 12 | OPRS1 | 9515 2402 | 196 | 7.038 | 0.2892 | Yes | ||
| 13 | PAICS | 16820 | 249 | 6.605 | 0.3074 | Yes | ||
| 14 | ELOVL4 | 19048 | 352 | 5.904 | 0.3207 | Yes | ||
| 15 | POLR3E | 18100 | 403 | 5.495 | 0.3355 | Yes | ||
| 16 | G6PC3 | 1221 20645 | 405 | 5.484 | 0.3529 | Yes | ||
| 17 | MORF4L2 | 12118 | 457 | 5.277 | 0.3670 | Yes | ||
| 18 | EXOSC2 | 15044 | 469 | 5.241 | 0.3831 | Yes | ||
| 19 | FKBP11 | 22136 | 511 | 4.996 | 0.3968 | Yes | ||
| 20 | ABCB6 | 13162 | 518 | 4.972 | 0.4123 | Yes | ||
| 21 | EIF4G1 | 22818 | 582 | 4.708 | 0.4239 | Yes | ||
| 22 | POLR2H | 10888 | 611 | 4.591 | 0.4370 | Yes | ||
| 23 | PPARGC1B | 9318 | 687 | 4.318 | 0.4467 | Yes | ||
| 24 | TIMM10 | 11390 2158 | 690 | 4.300 | 0.4603 | Yes | ||
| 25 | HYAL2 | 4891 | 855 | 3.806 | 0.4635 | Yes | ||
| 26 | PFDN2 | 9551 | 989 | 3.350 | 0.4669 | Yes | ||
| 27 | ACVR2B | 19276 | 1034 | 3.224 | 0.4748 | Yes | ||
| 28 | RPS19 | 5398 | 1114 | 3.041 | 0.4802 | Yes | ||
| 29 | ALDH3B1 | 12569 23949 | 1142 | 2.990 | 0.4883 | Yes | ||
| 30 | DIABLO | 16375 | 1230 | 2.812 | 0.4925 | Yes | ||
| 31 | ENO3 | 8905 | 1366 | 2.485 | 0.4931 | Yes | ||
| 32 | SH3KBP1 | 2631 12206 7189 | 1557 | 2.182 | 0.4898 | Yes | ||
| 33 | HSPH1 | 16282 | 1581 | 2.147 | 0.4954 | Yes | ||
| 34 | MYOHD1 | 20724 | 1691 | 1.991 | 0.4958 | Yes | ||
| 35 | SLC20A1 | 14855 | 1703 | 1.969 | 0.5015 | Yes | ||
| 36 | STMN1 | 9261 | 1819 | 1.798 | 0.5010 | Yes | ||
| 37 | PSMB3 | 11180 | 1849 | 1.748 | 0.5050 | Yes | ||
| 38 | EIF4B | 13279 7979 | 1871 | 1.716 | 0.5093 | Yes | ||
| 39 | SUCLG2 | 17062 5545 | 1920 | 1.646 | 0.5119 | Yes | ||
| 40 | PABPC4 | 10510 2418 2548 | 1938 | 1.608 | 0.5161 | Yes | ||
| 41 | PABPC1 | 5219 9522 9523 23572 | 1969 | 1.581 | 0.5196 | Yes | ||
| 42 | RHEBL1 | 12781 | 2144 | 1.385 | 0.5145 | No | ||
| 43 | HMOX1 | 18565 | 2213 | 1.291 | 0.5150 | No | ||
| 44 | SRP72 | 16819 | 2354 | 1.113 | 0.5109 | No | ||
| 45 | CENTB5 | 15961 | 2385 | 1.076 | 0.5127 | No | ||
| 46 | NLN | 3234 21358 | 2399 | 1.064 | 0.5154 | No | ||
| 47 | MPP3 | 8853 4630 5334 | 2534 | 0.982 | 0.5112 | No | ||
| 48 | NR1D1 | 4650 20258 949 | 2687 | 0.830 | 0.5056 | No | ||
| 49 | HSPBAP1 | 7321 | 2701 | 0.820 | 0.5076 | No | ||
| 50 | IVNS1ABP | 4384 4050 | 2745 | 0.769 | 0.5077 | No | ||
| 51 | CRMP1 | 16860 | 2782 | 0.740 | 0.5081 | No | ||
| 52 | SYT3 | 18264 | 2831 | 0.704 | 0.5077 | No | ||
| 53 | SLC39A7 | 23022 | 2832 | 0.703 | 0.5100 | No | ||
| 54 | TIMM9 | 19686 | 2835 | 0.698 | 0.5121 | No | ||
| 55 | IPO13 | 15784 | 2844 | 0.682 | 0.5138 | No | ||
| 56 | UBE1 | 24370 2551 | 2863 | 0.668 | 0.5150 | No | ||
| 57 | AVP | 14428 | 2994 | 0.560 | 0.5097 | No | ||
| 58 | RPL13A | 10226 79 | 3156 | 0.459 | 0.5024 | No | ||
| 59 | WEE1 | 18127 | 3229 | 0.429 | 0.4999 | No | ||
| 60 | FARP1 | 5849 10279 | 3392 | 0.355 | 0.4922 | No | ||
| 61 | ADCY3 | 21329 894 | 3959 | 0.220 | 0.4622 | No | ||
| 62 | GPRC5C | 12864 | 4033 | 0.209 | 0.4589 | No | ||
| 63 | ADAMTS17 | 598 | 4054 | 0.206 | 0.4585 | No | ||
| 64 | GTF2H1 | 4069 18236 | 4080 | 0.203 | 0.4578 | No | ||
| 65 | XRN2 | 2936 6302 2905 | 4224 | 0.182 | 0.4506 | No | ||
| 66 | BCKDHA | 17925 | 4286 | 0.175 | 0.4479 | No | ||
| 67 | DAZAP1 | 19945 | 4342 | 0.168 | 0.4454 | No | ||
| 68 | TBX4 | 5633 | 4363 | 0.164 | 0.4448 | No | ||
| 69 | CLCN2 | 22637 | 4385 | 0.162 | 0.4442 | No | ||
| 70 | CHST11 | 19921 | 4464 | 0.154 | 0.4405 | No | ||
| 71 | SDC1 | 5560 9953 | 4550 | 0.146 | 0.4363 | No | ||
| 72 | TNFRSF21 | 8244 | 4660 | 0.135 | 0.4309 | No | ||
| 73 | SGTB | 21571 | 4825 | 0.123 | 0.4223 | No | ||
| 74 | SET | 7070 | 4881 | 0.120 | 0.4197 | No | ||
| 75 | LTBR | 16993 | 4882 | 0.120 | 0.4201 | No | ||
| 76 | RPL19 | 9736 | 4895 | 0.119 | 0.4199 | No | ||
| 77 | HOXC13 | 22344 | 5311 | 0.095 | 0.3976 | No | ||
| 78 | FLT3 | 16288 | 5395 | 0.091 | 0.3934 | No | ||
| 79 | CACNA1D | 4464 8675 | 5729 | 0.076 | 0.3756 | No | ||
| 80 | MYST3 | 18651 | 5861 | 0.071 | 0.3687 | No | ||
| 81 | NPM1 | 1196 | 5908 | 0.070 | 0.3664 | No | ||
| 82 | PLA2G4A | 13809 | 6106 | 0.063 | 0.3560 | No | ||
| 83 | AMPD2 | 1931 15197 | 6237 | 0.059 | 0.3491 | No | ||
| 84 | LHX9 | 9276 | 6325 | 0.057 | 0.3446 | No | ||
| 85 | EIF3S1 | 905 8114 | 6441 | 0.054 | 0.3385 | No | ||
| 86 | LRP8 | 16149 2425 | 6500 | 0.053 | 0.3355 | No | ||
| 87 | POGK | 7739 | 6674 | 0.049 | 0.3263 | No | ||
| 88 | SEMA7A | 19426 9803 | 6735 | 0.048 | 0.3232 | No | ||
| 89 | KIAA0427 | 11228 | 6740 | 0.048 | 0.3231 | No | ||
| 90 | SYT6 | 13437 7042 12040 | 6823 | 0.046 | 0.3188 | No | ||
| 91 | CTSF | 23772 | 7147 | 0.039 | 0.3014 | No | ||
| 92 | FADS3 | 7214 | 7226 | 0.038 | 0.2973 | No | ||
| 93 | FGF11 | 20383 | 7450 | 0.034 | 0.2853 | No | ||
| 94 | EIF4E | 15403 1827 8890 | 7583 | 0.031 | 0.2782 | No | ||
| 95 | HOXC12 | 22343 | 7684 | 0.030 | 0.2729 | No | ||
| 96 | HSPA4 | 1411 20465 | 7711 | 0.029 | 0.2716 | No | ||
| 97 | DAZL | 22944 | 7826 | 0.027 | 0.2655 | No | ||
| 98 | ARX | 24290 | 7839 | 0.027 | 0.2649 | No | ||
| 99 | SNCAIP | 12589 | 7868 | 0.027 | 0.2635 | No | ||
| 100 | DRD1 | 4638 | 7875 | 0.027 | 0.2633 | No | ||
| 101 | PRKCE | 9575 | 7915 | 0.026 | 0.2612 | No | ||
| 102 | TDRD1 | 13510 23818 | 7957 | 0.026 | 0.2591 | No | ||
| 103 | MNT | 20784 | 8098 | 0.023 | 0.2516 | No | ||
| 104 | SLC12A5 | 14731 | 8596 | 0.016 | 0.2247 | No | ||
| 105 | NKX2-3 | 5177 23845 | 8761 | 0.015 | 0.2158 | No | ||
| 106 | CGN | 12896 12897 7701 | 8869 | 0.013 | 0.2100 | No | ||
| 107 | MTHFD1 | 2132 21238 | 8889 | 0.013 | 0.2091 | No | ||
| 108 | HOXA3 | 9108 1075 | 9000 | 0.012 | 0.2031 | No | ||
| 109 | FGF14 | 21716 3005 3463 | 9073 | 0.011 | 0.1992 | No | ||
| 110 | RORC | 9732 | 9122 | 0.010 | 0.1967 | No | ||
| 111 | SLC6A15 | 8335 3321 | 9209 | 0.009 | 0.1920 | No | ||
| 112 | PAX6 | 5223 | 9241 | 0.009 | 0.1904 | No | ||
| 113 | NXPH1 | 17524 | 9695 | 0.003 | 0.1658 | No | ||
| 114 | COL2A1 | 22145 4542 | 9823 | 0.001 | 0.1589 | No | ||
| 115 | CHRM1 | 23943 | 9991 | -0.001 | 0.1499 | No | ||
| 116 | TLL1 | 10188 | 9995 | -0.001 | 0.1497 | No | ||
| 117 | SCFD2 | 5607 | 9997 | -0.001 | 0.1497 | No | ||
| 118 | GPD1 | 22361 | 10004 | -0.001 | 0.1493 | No | ||
| 119 | NPTX1 | 20126 | 10028 | -0.001 | 0.1481 | No | ||
| 120 | SLC26A2 | 23427 | 10489 | -0.007 | 0.1232 | No | ||
| 121 | USP2 | 19480 3052 3043 | 10584 | -0.008 | 0.1181 | No | ||
| 122 | HOXA4 | 17148 | 10724 | -0.010 | 0.1106 | No | ||
| 123 | HPCA | 9118 | 10928 | -0.013 | 0.0996 | No | ||
| 124 | GTF2A1 | 2064 8187 8188 13512 | 10949 | -0.013 | 0.0986 | No | ||
| 125 | HHIP | 4849 | 11477 | -0.021 | 0.0700 | No | ||
| 126 | CNNM1 | 23846 | 11566 | -0.023 | 0.0653 | No | ||
| 127 | TIAL1 | 996 5753 | 11790 | -0.026 | 0.0533 | No | ||
| 128 | PIGW | 20312 | 11815 | -0.027 | 0.0521 | No | ||
| 129 | HS3ST3A1 | 20844 | 11888 | -0.028 | 0.0483 | No | ||
| 130 | UBXD3 | 15701 | 11966 | -0.029 | 0.0442 | No | ||
| 131 | SC5DL | 6165 | 11980 | -0.030 | 0.0436 | No | ||
| 132 | DNMT3A | 2167 21330 | 12095 | -0.032 | 0.0375 | No | ||
| 133 | SATB2 | 5604 | 12242 | -0.035 | 0.0297 | No | ||
| 134 | ABCA1 | 15881 | 12409 | -0.039 | 0.0208 | No | ||
| 135 | RUNX2 | 4480 8700 | 12564 | -0.043 | 0.0126 | No | ||
| 136 | DNAJB9 | 6565 | 12984 | -0.054 | -0.0100 | No | ||
| 137 | LIN28 | 15723 | 13081 | -0.057 | -0.0150 | No | ||
| 138 | GRIN2A | 22668 | 13281 | -0.064 | -0.0256 | No | ||
| 139 | HPCAL4 | 16096 | 13476 | -0.072 | -0.0359 | No | ||
| 140 | MCM8 | 14834 | 13711 | -0.084 | -0.0483 | No | ||
| 141 | CAMK4 | 4473 | 13964 | -0.100 | -0.0617 | No | ||
| 142 | PER1 | 20827 | 14194 | -0.119 | -0.0737 | No | ||
| 143 | ENPP6 | 6816 11583 | 14254 | -0.125 | -0.0765 | No | ||
| 144 | LAP3 | 12442 | 14326 | -0.132 | -0.0800 | No | ||
| 145 | SOCS5 | 1619 23141 | 14532 | -0.155 | -0.0906 | No | ||
| 146 | RUSC1 | 1853 15283 | 14848 | -0.209 | -0.1070 | No | ||
| 147 | GRK6 | 6449 | 15024 | -0.244 | -0.1157 | No | ||
| 148 | PDK2 | 20285 | 15084 | -0.259 | -0.1181 | No | ||
| 149 | PTMA | 9657 | 15118 | -0.268 | -0.1190 | No | ||
| 150 | B4GALT2 | 11990 | 15125 | -0.270 | -0.1185 | No | ||
| 151 | HMGN2 | 9095 | 15229 | -0.297 | -0.1231 | No | ||
| 152 | RPL22 | 15979 | 15394 | -0.349 | -0.1309 | No | ||
| 153 | RXRB | 23285 9768 | 15412 | -0.356 | -0.1307 | No | ||
| 154 | ETV1 | 4688 8925 8924 | 15473 | -0.378 | -0.1328 | No | ||
| 155 | ETV4 | 20212 | 15513 | -0.403 | -0.1336 | No | ||
| 156 | SNX5 | 12783 14410 | 15667 | -0.474 | -0.1404 | No | ||
| 157 | BMP2K | 3657 16786 | 15733 | -0.510 | -0.1423 | No | ||
| 158 | SLC31A2 | 9827 | 16027 | -0.750 | -0.1558 | No | ||
| 159 | ARF6 | 21252 | 16303 | -1.004 | -0.1675 | No | ||
| 160 | EFNB1 | 24285 | 16362 | -1.056 | -0.1673 | No | ||
| 161 | PFN1 | 9555 | 16390 | -1.090 | -0.1653 | No | ||
| 162 | MAX | 21034 | 16454 | -1.158 | -0.1650 | No | ||
| 163 | FCHSD2 | 18172 | 16709 | -1.525 | -0.1739 | No | ||
| 164 | FHOD1 | 18772 | 16792 | -1.656 | -0.1731 | No | ||
| 165 | TEF | 2287 22414 2192 | 16862 | -1.729 | -0.1713 | No | ||
| 166 | AKAP12 | 17600 | 17015 | -1.927 | -0.1734 | No | ||
| 167 | ANXA6 | 20449 | 17024 | -1.936 | -0.1677 | No | ||
| 168 | GPX1 | 19310 | 17165 | -2.181 | -0.1684 | No | ||
| 169 | SIRT1 | 19745 | 17277 | -2.379 | -0.1668 | No | ||
| 170 | MAT2A | 10553 10554 6099 | 17281 | -2.385 | -0.1594 | No | ||
| 171 | CDC14A | 15184 | 17350 | -2.556 | -0.1549 | No | ||
| 172 | MXD4 | 9346 | 17505 | -2.876 | -0.1541 | No | ||
| 173 | EIF2C2 | 6247 | 17620 | -3.135 | -0.1503 | No | ||
| 174 | PSEN2 | 13736 | 17666 | -3.289 | -0.1422 | No | ||
| 175 | BCAS3 | 5314 | 17668 | -3.294 | -0.1318 | No | ||
| 176 | GNA13 | 20617 | 17715 | -3.402 | -0.1235 | No | ||
| 177 | BCL9L | 19475 | 17915 | -3.935 | -0.1217 | No | ||
| 178 | BMP4 | 21863 | 17960 | -4.096 | -0.1111 | No | ||
| 179 | ARRDC3 | 4191 | 18030 | -4.350 | -0.1009 | No | ||
| 180 | GABARAP | 20815 | 18114 | -4.664 | -0.0906 | No | ||
| 181 | H3F3A | 4838 | 18120 | -4.694 | -0.0759 | No | ||
| 182 | WBP2 | 20146 | 18156 | -4.817 | -0.0624 | No | ||
| 183 | PPP1R9B | 20698 | 18242 | -5.266 | -0.0503 | No | ||
| 184 | HERPUD1 | 18522 | 18388 | -6.080 | -0.0388 | No | ||
| 185 | BHLHB2 | 17340 | 18498 | -7.262 | -0.0215 | No | ||
| 186 | BCL6 | 22624 | 18580 | -8.747 | 0.0020 | No |