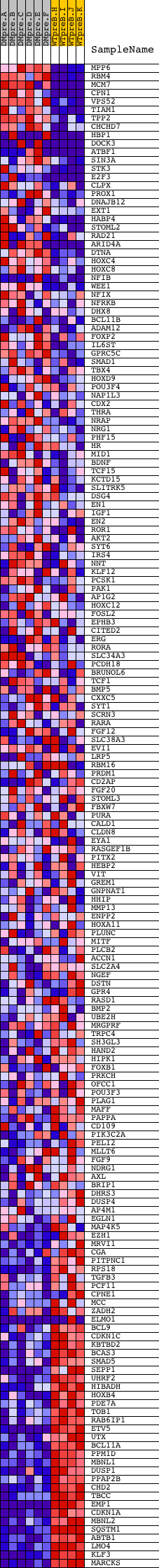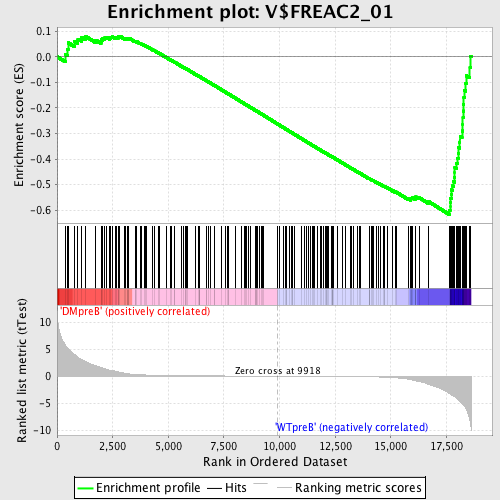
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
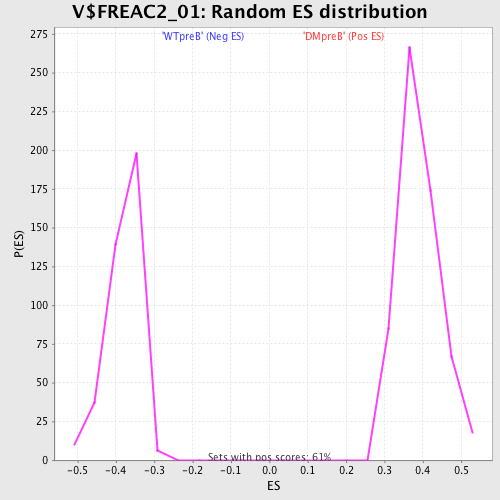
| GeneSet | V$FREAC2_01 |
| Enrichment Score (ES) | -0.61637294 |
| Normalized Enrichment Score (NES) | -1.6334445 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.050572433 |
| FWER p-Value | 0.189 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MPP6 | 17448 | 371 | 5.712 | 0.0090 | No | ||
| 2 | RBM4 | 9706 3693 | 476 | 5.199 | 0.0298 | No | ||
| 3 | MCM7 | 9372 3568 | 497 | 5.089 | 0.0546 | No | ||
| 4 | CPN1 | 23667 | 790 | 3.964 | 0.0589 | No | ||
| 5 | VPS52 | 23287 1622 | 935 | 3.524 | 0.0691 | No | ||
| 6 | TIAM1 | 22547 | 1097 | 3.076 | 0.0760 | No | ||
| 7 | TPP2 | 14257 | 1289 | 2.666 | 0.0792 | No | ||
| 8 | CHCHD7 | 16280 | 1731 | 1.922 | 0.0651 | No | ||
| 9 | HBP1 | 2048 2162 21086 | 1975 | 1.578 | 0.0599 | No | ||
| 10 | DOCK3 | 9920 5537 10751 | 1983 | 1.574 | 0.0675 | No | ||
| 11 | ATBF1 | 916 8633 | 2048 | 1.498 | 0.0717 | No | ||
| 12 | SIN3A | 5442 | 2148 | 1.383 | 0.0734 | No | ||
| 13 | STK3 | 7084 | 2207 | 1.307 | 0.0769 | No | ||
| 14 | E2F3 | 21500 8874 | 2367 | 1.098 | 0.0738 | No | ||
| 15 | CLPX | 19403 | 2421 | 1.049 | 0.0763 | No | ||
| 16 | PROX1 | 9623 | 2473 | 1.006 | 0.0787 | No | ||
| 17 | DNAJB12 | 7146 | 2637 | 0.882 | 0.0743 | No | ||
| 18 | EXT1 | 22297 | 2669 | 0.843 | 0.0769 | No | ||
| 19 | HABP4 | 12145 | 2744 | 0.771 | 0.0768 | No | ||
| 20 | STOML2 | 15902 | 2765 | 0.752 | 0.0796 | No | ||
| 21 | RAD21 | 22298 | 2816 | 0.716 | 0.0805 | No | ||
| 22 | ARID4A | 2080 6215 | 3041 | 0.533 | 0.0711 | No | ||
| 23 | DTNA | 8870 4647 23611 1991 | 3063 | 0.521 | 0.0726 | No | ||
| 24 | HOXC4 | 4864 | 3152 | 0.460 | 0.0701 | No | ||
| 25 | HOXC8 | 4865 | 3177 | 0.451 | 0.0711 | No | ||
| 26 | NFIB | 15855 | 3226 | 0.430 | 0.0707 | No | ||
| 27 | WEE1 | 18127 | 3229 | 0.429 | 0.0728 | No | ||
| 28 | NFIX | 9458 | 3501 | 0.326 | 0.0598 | No | ||
| 29 | NFRKB | 10642 | 3521 | 0.321 | 0.0604 | No | ||
| 30 | DHX8 | 20649 | 3583 | 0.306 | 0.0586 | No | ||
| 31 | BCL11B | 7190 | 3746 | 0.263 | 0.0512 | No | ||
| 32 | ADAM12 | 2207 8545 | 3812 | 0.249 | 0.0489 | No | ||
| 33 | FOXP2 | 4312 17523 8523 | 3935 | 0.225 | 0.0434 | No | ||
| 34 | IL6ST | 4920 | 3953 | 0.221 | 0.0436 | No | ||
| 35 | GPRC5C | 12864 | 4033 | 0.209 | 0.0404 | No | ||
| 36 | SMAD1 | 18831 922 | 4300 | 0.173 | 0.0269 | No | ||
| 37 | TBX4 | 5633 | 4363 | 0.164 | 0.0243 | No | ||
| 38 | HOXD9 | 14982 | 4547 | 0.146 | 0.0152 | No | ||
| 39 | POU3F4 | 347 | 4618 | 0.139 | 0.0121 | No | ||
| 40 | NAP1L3 | 24266 | 4913 | 0.117 | -0.0033 | No | ||
| 41 | CDX2 | 16289 | 5116 | 0.106 | -0.0137 | No | ||
| 42 | THRA | 1447 10171 1406 | 5121 | 0.106 | -0.0134 | No | ||
| 43 | NRAP | 23642 | 5283 | 0.097 | -0.0216 | No | ||
| 44 | NRG1 | 3885 | 5288 | 0.097 | -0.0213 | No | ||
| 45 | PHF15 | 20467 8050 | 5571 | 0.083 | -0.0362 | No | ||
| 46 | HR | 3622 9119 | 5698 | 0.077 | -0.0427 | No | ||
| 47 | MID1 | 5097 5098 | 5763 | 0.074 | -0.0458 | No | ||
| 48 | BDNF | 14926 2797 | 5766 | 0.074 | -0.0455 | No | ||
| 49 | TCF15 | 14798 | 5807 | 0.072 | -0.0473 | No | ||
| 50 | KCTD15 | 17864 | 5849 | 0.071 | -0.0491 | No | ||
| 51 | SLITRK5 | 21939 | 6241 | 0.059 | -0.0700 | No | ||
| 52 | DSG4 | 9262 23616 | 6374 | 0.056 | -0.0769 | No | ||
| 53 | EN1 | 387 14155 | 6404 | 0.055 | -0.0782 | No | ||
| 54 | IGF1 | 3352 9156 3409 | 6411 | 0.055 | -0.0783 | No | ||
| 55 | EN2 | 16898 | 6421 | 0.055 | -0.0785 | No | ||
| 56 | ROR1 | 6472 | 6713 | 0.049 | -0.0940 | No | ||
| 57 | AKT2 | 4365 4366 | 6728 | 0.048 | -0.0945 | No | ||
| 58 | SYT6 | 13437 7042 12040 | 6823 | 0.046 | -0.0994 | No | ||
| 59 | IRS4 | 9183 4926 | 6907 | 0.044 | -0.1037 | No | ||
| 60 | NNT | 3273 5181 9471 3238 | 7053 | 0.041 | -0.1113 | No | ||
| 61 | KLF12 | 4960 2637 21732 9228 | 7086 | 0.040 | -0.1128 | No | ||
| 62 | PCSK1 | 21600 | 7368 | 0.035 | -0.1279 | No | ||
| 63 | PAK1 | 9527 | 7590 | 0.031 | -0.1397 | No | ||
| 64 | AP1G2 | 2677 21827 | 7659 | 0.030 | -0.1433 | No | ||
| 65 | HOXC12 | 22343 | 7684 | 0.030 | -0.1444 | No | ||
| 66 | FOSL2 | 4733 8978 16878 | 7715 | 0.029 | -0.1459 | No | ||
| 67 | EPHB3 | 22816 | 8008 | 0.025 | -0.1616 | No | ||
| 68 | CITED2 | 5118 14477 | 8285 | 0.021 | -0.1765 | No | ||
| 69 | ERG | 1686 8915 | 8403 | 0.019 | -0.1827 | No | ||
| 70 | RORA | 3019 5388 | 8471 | 0.018 | -0.1863 | No | ||
| 71 | SLC34A3 | 14664 | 8517 | 0.018 | -0.1886 | No | ||
| 72 | PCDH18 | 15348 | 8590 | 0.017 | -0.1924 | No | ||
| 73 | BRUNOL6 | 19421 | 8706 | 0.015 | -0.1986 | No | ||
| 74 | TCF1 | 16416 | 8918 | 0.013 | -0.2100 | No | ||
| 75 | BMP5 | 19376 | 8936 | 0.012 | -0.2108 | No | ||
| 76 | CXXC5 | 12523 | 8946 | 0.012 | -0.2113 | No | ||
| 77 | SYT1 | 5565 | 8949 | 0.012 | -0.2113 | No | ||
| 78 | SCRN3 | 14984 | 9003 | 0.012 | -0.2141 | No | ||
| 79 | RARA | 5358 | 9085 | 0.011 | -0.2185 | No | ||
| 80 | FGF12 | 1723 22621 | 9206 | 0.009 | -0.2249 | No | ||
| 81 | SLC38A3 | 13319 | 9233 | 0.009 | -0.2263 | No | ||
| 82 | EVI1 | 4689 8926 | 9266 | 0.008 | -0.2280 | No | ||
| 83 | LRP5 | 23948 9285 | 9907 | 0.000 | -0.2627 | No | ||
| 84 | RBM16 | 11492 8387 | 9923 | -0.000 | -0.2635 | No | ||
| 85 | PRDM1 | 19775 3337 | 9987 | -0.001 | -0.2669 | No | ||
| 86 | CD2AP | 22975 | 10173 | -0.003 | -0.2769 | No | ||
| 87 | FGF20 | 18892 | 10245 | -0.004 | -0.2808 | No | ||
| 88 | STOML3 | 15594 | 10304 | -0.005 | -0.2839 | No | ||
| 89 | FBXW7 | 1805 11928 | 10321 | -0.005 | -0.2847 | No | ||
| 90 | PURA | 9670 | 10425 | -0.006 | -0.2903 | No | ||
| 91 | CALD1 | 4273 8463 | 10526 | -0.008 | -0.2957 | No | ||
| 92 | CLDN8 | 22549 1747 | 10573 | -0.008 | -0.2981 | No | ||
| 93 | EYA1 | 4695 4061 | 10600 | -0.009 | -0.2995 | No | ||
| 94 | RASGEF1B | 11515 16469 | 10670 | -0.010 | -0.3032 | No | ||
| 95 | PITX2 | 15424 1878 | 10992 | -0.014 | -0.3205 | No | ||
| 96 | HEBP2 | 19812 | 11112 | -0.016 | -0.3269 | No | ||
| 97 | VIT | 23155 | 11187 | -0.017 | -0.3308 | No | ||
| 98 | GREM1 | 14485 | 11304 | -0.019 | -0.3370 | No | ||
| 99 | GNPNAT1 | 12027 7031 | 11410 | -0.020 | -0.3426 | No | ||
| 100 | HHIP | 4849 | 11477 | -0.021 | -0.3461 | No | ||
| 101 | MMP13 | 19573 3007 | 11511 | -0.022 | -0.3478 | No | ||
| 102 | ENPP2 | 9548 | 11567 | -0.023 | -0.3506 | No | ||
| 103 | HOXA11 | 17146 | 11692 | -0.025 | -0.3572 | No | ||
| 104 | PLUNC | 9593 | 11833 | -0.027 | -0.3647 | No | ||
| 105 | MITF | 17349 | 11903 | -0.028 | -0.3683 | No | ||
| 106 | PLCB2 | 5262 | 11963 | -0.029 | -0.3713 | No | ||
| 107 | ACCN1 | 20327 1194 | 12053 | -0.031 | -0.3760 | No | ||
| 108 | SLC2A4 | 20380 | 12102 | -0.032 | -0.3784 | No | ||
| 109 | NGEF | 13891 | 12109 | -0.032 | -0.3786 | No | ||
| 110 | DSTN | 14823 | 12168 | -0.034 | -0.3816 | No | ||
| 111 | GPR4 | 18364 | 12216 | -0.034 | -0.3840 | No | ||
| 112 | RASD1 | 20424 | 12352 | -0.038 | -0.3911 | No | ||
| 113 | BMP2 | 14833 | 12361 | -0.038 | -0.3913 | No | ||
| 114 | UBE2H | 5823 5822 | 12376 | -0.038 | -0.3919 | No | ||
| 115 | MRGPRF | 17995 | 12378 | -0.038 | -0.3918 | No | ||
| 116 | TRPC4 | 15593 | 12401 | -0.039 | -0.3927 | No | ||
| 117 | SH3GL3 | 18201 | 12587 | -0.044 | -0.4026 | No | ||
| 118 | HAND2 | 18615 | 12807 | -0.049 | -0.4142 | No | ||
| 119 | HIPK1 | 4851 | 12960 | -0.053 | -0.4222 | No | ||
| 120 | FOXB1 | 12254 19069 | 13190 | -0.061 | -0.4343 | No | ||
| 121 | PRKCH | 21246 | 13227 | -0.062 | -0.4359 | No | ||
| 122 | OFCC1 | 21486 | 13329 | -0.066 | -0.4411 | No | ||
| 123 | POU3F3 | 14261 | 13516 | -0.073 | -0.4508 | No | ||
| 124 | PLAG1 | 7148 15949 12161 | 13589 | -0.078 | -0.4543 | No | ||
| 125 | MAFF | 22423 2243 | 13615 | -0.079 | -0.4552 | No | ||
| 126 | PAPPA | 16190 | 14030 | -0.104 | -0.4772 | No | ||
| 127 | CD109 | 6174 10656 19369 | 14038 | -0.105 | -0.4770 | No | ||
| 128 | PIK3C2A | 17667 | 14052 | -0.106 | -0.4772 | No | ||
| 129 | PELI2 | 8217 | 14133 | -0.112 | -0.4809 | No | ||
| 130 | MLLT6 | 20678 | 14163 | -0.115 | -0.4819 | No | ||
| 131 | FGF9 | 8966 | 14222 | -0.121 | -0.4845 | No | ||
| 132 | NDRG1 | 22276 | 14367 | -0.137 | -0.4916 | No | ||
| 133 | AXL | 17922 | 14463 | -0.148 | -0.4960 | No | ||
| 134 | BRIP1 | 20311 | 14537 | -0.156 | -0.4991 | No | ||
| 135 | DHRS3 | 15997 | 14663 | -0.177 | -0.5050 | No | ||
| 136 | DUSP4 | 18632 3820 | 14695 | -0.182 | -0.5058 | No | ||
| 137 | AP4M1 | 3499 16656 | 14702 | -0.183 | -0.5052 | No | ||
| 138 | EGLN1 | 8504 18707 4298 | 14854 | -0.211 | -0.5123 | No | ||
| 139 | MAP4K5 | 11853 21048 2101 2142 | 15096 | -0.263 | -0.5240 | No | ||
| 140 | EZH1 | 20217 | 15194 | -0.287 | -0.5278 | No | ||
| 141 | MRVI1 | 17674 1020 2526 | 15236 | -0.298 | -0.5285 | No | ||
| 142 | CGA | 16246 | 15780 | -0.538 | -0.5552 | No | ||
| 143 | PITPNC1 | 7759 | 15898 | -0.634 | -0.5583 | No | ||
| 144 | RPS18 | 1529 5397 1603 | 15907 | -0.640 | -0.5555 | No | ||
| 145 | TGFB3 | 10161 | 15913 | -0.649 | -0.5525 | No | ||
| 146 | PCF11 | 121 | 15957 | -0.683 | -0.5513 | No | ||
| 147 | CPNE1 | 11186 | 16094 | -0.816 | -0.5546 | No | ||
| 148 | MCC | 552 | 16095 | -0.817 | -0.5504 | No | ||
| 149 | ZADH2 | 23501 | 16127 | -0.854 | -0.5477 | No | ||
| 150 | ELMO1 | 3275 8939 3202 3271 3254 3235 3164 3175 3274 3188 3293 3214 3218 3239 | 16273 | -0.995 | -0.5505 | No | ||
| 151 | BCL9 | 15232 | 16684 | -1.483 | -0.5652 | No | ||
| 152 | CDKN1C | 17546 | 17628 | -3.172 | -0.6002 | Yes | ||
| 153 | KBTBD2 | 17137 | 17665 | -3.280 | -0.5855 | Yes | ||
| 154 | BCAS3 | 5314 | 17668 | -3.294 | -0.5688 | Yes | ||
| 155 | SMAD5 | 21621 | 17677 | -3.317 | -0.5524 | Yes | ||
| 156 | SEPP1 | 22524 | 17720 | -3.429 | -0.5372 | Yes | ||
| 157 | UHRF2 | 3731 4262 8452 23887 | 17733 | -3.460 | -0.5202 | Yes | ||
| 158 | HIBADH | 17144 | 17782 | -3.569 | -0.5047 | Yes | ||
| 159 | HOXB4 | 20686 | 17827 | -3.678 | -0.4883 | Yes | ||
| 160 | PDE7A | 9544 1788 | 17861 | -3.771 | -0.4709 | Yes | ||
| 161 | TOB1 | 20703 | 17875 | -3.812 | -0.4522 | Yes | ||
| 162 | RAB6IP1 | 17676 | 17879 | -3.826 | -0.4329 | Yes | ||
| 163 | ETV5 | 22630 | 17951 | -4.075 | -0.4160 | Yes | ||
| 164 | UTX | 10266 2574 | 17994 | -4.204 | -0.3969 | Yes | ||
| 165 | BCL11A | 4691 | 18039 | -4.409 | -0.3769 | Yes | ||
| 166 | PPM1D | 20721 | 18053 | -4.474 | -0.3548 | Yes | ||
| 167 | MBNL1 | 1921 15582 | 18100 | -4.631 | -0.3337 | Yes | ||
| 168 | DUSP1 | 23061 | 18109 | -4.651 | -0.3105 | Yes | ||
| 169 | PPAP2B | 7503 16161 | 18203 | -5.029 | -0.2899 | Yes | ||
| 170 | CHD2 | 10847 | 18237 | -5.235 | -0.2651 | Yes | ||
| 171 | TBCC | 23215 | 18243 | -5.273 | -0.2385 | Yes | ||
| 172 | EMP1 | 17260 | 18255 | -5.322 | -0.2120 | Yes | ||
| 173 | CDKN1A | 4511 8729 | 18269 | -5.369 | -0.1854 | Yes | ||
| 174 | MBNL2 | 21932 | 18284 | -5.431 | -0.1585 | Yes | ||
| 175 | SQSTM1 | 9517 | 18310 | -5.605 | -0.1313 | Yes | ||
| 176 | ABTB1 | 17075 | 18369 | -5.928 | -0.1043 | Yes | ||
| 177 | LMO4 | 15151 | 18404 | -6.210 | -0.0745 | Yes | ||
| 178 | KLF3 | 4961 | 18557 | -8.119 | -0.0414 | Yes | ||
| 179 | MARCKS | 9331 | 18581 | -8.761 | 0.0019 | Yes |