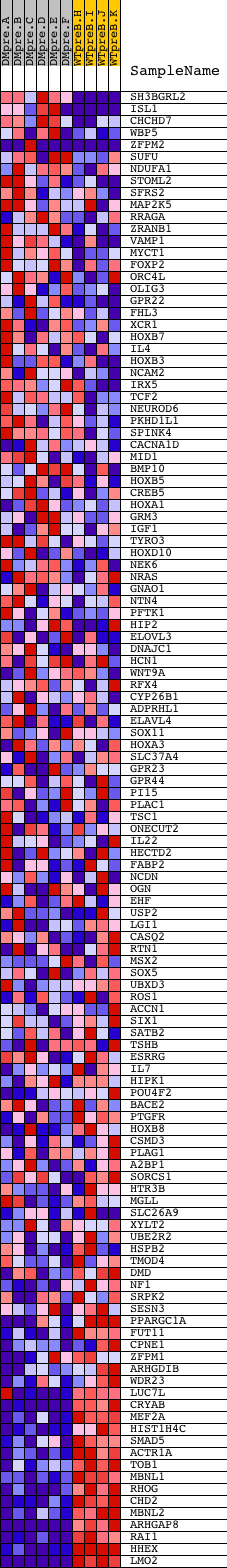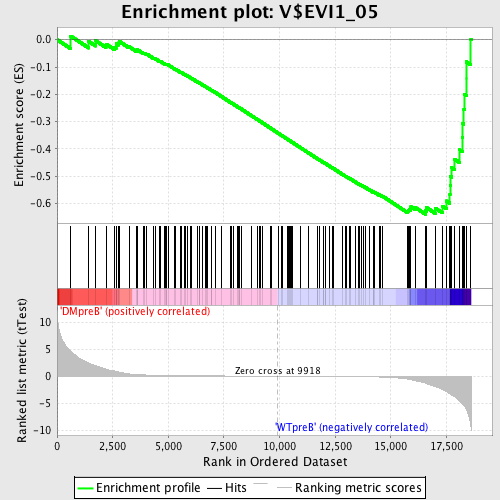
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | V$EVI1_05 |
| Enrichment Score (ES) | -0.6400518 |
| Normalized Enrichment Score (NES) | -1.6158867 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.055623066 |
| FWER p-Value | 0.247 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SH3BGRL2 | 5601 | 613 | 4.586 | 0.0131 | No | ||
| 2 | ISL1 | 21338 | 1420 | 2.387 | -0.0064 | No | ||
| 3 | CHCHD7 | 16280 | 1731 | 1.922 | -0.0038 | No | ||
| 4 | WBP5 | 10297 | 2199 | 1.314 | -0.0158 | No | ||
| 5 | ZFPM2 | 22483 | 2596 | 0.926 | -0.0279 | No | ||
| 6 | SUFU | 6283 | 2651 | 0.860 | -0.0221 | No | ||
| 7 | NDUFA1 | 24170 | 2662 | 0.846 | -0.0141 | No | ||
| 8 | STOML2 | 15902 | 2765 | 0.752 | -0.0120 | No | ||
| 9 | SFRS2 | 9807 20136 | 2789 | 0.733 | -0.0059 | No | ||
| 10 | MAP2K5 | 19088 | 3237 | 0.426 | -0.0258 | No | ||
| 11 | RRAGA | 16180 | 3552 | 0.313 | -0.0396 | No | ||
| 12 | ZRANB1 | 6884 11702 | 3600 | 0.302 | -0.0391 | No | ||
| 13 | VAMP1 | 5846 1043 | 3620 | 0.296 | -0.0371 | No | ||
| 14 | MYCT1 | 7572 | 3893 | 0.231 | -0.0495 | No | ||
| 15 | FOXP2 | 4312 17523 8523 | 3935 | 0.225 | -0.0494 | No | ||
| 16 | ORC4L | 11172 6460 | 4035 | 0.208 | -0.0527 | No | ||
| 17 | OLIG3 | 20084 | 4338 | 0.168 | -0.0673 | No | ||
| 18 | GPR22 | 21087 | 4421 | 0.158 | -0.0702 | No | ||
| 19 | FHL3 | 16092 2457 | 4613 | 0.139 | -0.0791 | No | ||
| 20 | XCR1 | 18953 | 4639 | 0.137 | -0.0790 | No | ||
| 21 | HOXB7 | 666 9110 | 4836 | 0.122 | -0.0884 | No | ||
| 22 | IL4 | 9174 | 4886 | 0.119 | -0.0899 | No | ||
| 23 | HOXB3 | 20685 | 4909 | 0.118 | -0.0899 | No | ||
| 24 | NCAM2 | 9445 5150 | 4929 | 0.117 | -0.0897 | No | ||
| 25 | IRX5 | 18527 | 4997 | 0.112 | -0.0922 | No | ||
| 26 | TCF2 | 1395 20727 | 5292 | 0.096 | -0.1071 | No | ||
| 27 | NEUROD6 | 17138 | 5317 | 0.095 | -0.1075 | No | ||
| 28 | PKHD1L1 | 22481 9650 | 5543 | 0.084 | -0.1188 | No | ||
| 29 | SPINK4 | 16241 | 5574 | 0.083 | -0.1196 | No | ||
| 30 | CACNA1D | 4464 8675 | 5729 | 0.076 | -0.1271 | No | ||
| 31 | MID1 | 5097 5098 | 5763 | 0.074 | -0.1282 | No | ||
| 32 | BMP10 | 17376 | 5864 | 0.071 | -0.1329 | No | ||
| 33 | HOXB5 | 20687 | 5990 | 0.067 | -0.1390 | No | ||
| 34 | CREB5 | 10551 | 6052 | 0.065 | -0.1416 | No | ||
| 35 | HOXA1 | 1005 17152 | 6290 | 0.058 | -0.1538 | No | ||
| 36 | GRM3 | 16929 | 6379 | 0.056 | -0.1580 | No | ||
| 37 | IGF1 | 3352 9156 3409 | 6411 | 0.055 | -0.1591 | No | ||
| 38 | TYRO3 | 5811 | 6547 | 0.052 | -0.1659 | No | ||
| 39 | HOXD10 | 14983 | 6657 | 0.050 | -0.1713 | No | ||
| 40 | NEK6 | 15018 | 6719 | 0.048 | -0.1741 | No | ||
| 41 | NRAS | 5191 | 6779 | 0.047 | -0.1768 | No | ||
| 42 | GNAO1 | 4785 3829 | 6926 | 0.044 | -0.1843 | No | ||
| 43 | NTN4 | 19903 | 6934 | 0.044 | -0.1842 | No | ||
| 44 | PFTK1 | 16924 | 7117 | 0.040 | -0.1937 | No | ||
| 45 | HIP2 | 3626 3485 16838 | 7403 | 0.035 | -0.2087 | No | ||
| 46 | ELOVL3 | 23809 | 7809 | 0.028 | -0.2303 | No | ||
| 47 | DNAJC1 | 8854 | 7818 | 0.028 | -0.2305 | No | ||
| 48 | HCN1 | 21557 | 7933 | 0.026 | -0.2364 | No | ||
| 49 | WNT9A | 5709 | 8121 | 0.023 | -0.2463 | No | ||
| 50 | RFX4 | 7721 3410 | 8136 | 0.023 | -0.2468 | No | ||
| 51 | CYP26B1 | 17093 | 8191 | 0.022 | -0.2495 | No | ||
| 52 | ADPRHL1 | 18930 3863 | 8195 | 0.022 | -0.2495 | No | ||
| 53 | ELAVL4 | 15805 4889 9137 | 8276 | 0.021 | -0.2536 | No | ||
| 54 | SOX11 | 5477 | 8733 | 0.015 | -0.2781 | No | ||
| 55 | HOXA3 | 9108 1075 | 9000 | 0.012 | -0.2923 | No | ||
| 56 | SLC37A4 | 8994 | 9107 | 0.010 | -0.2980 | No | ||
| 57 | GPR23 | 13428 | 9157 | 0.010 | -0.3005 | No | ||
| 58 | GPR44 | 23926 | 9219 | 0.009 | -0.3037 | No | ||
| 59 | PI15 | 13586 | 9569 | 0.004 | -0.3226 | No | ||
| 60 | PLAC1 | 12079 | 9642 | 0.003 | -0.3264 | No | ||
| 61 | TSC1 | 7235 | 9972 | -0.001 | -0.3442 | No | ||
| 62 | ONECUT2 | 10346 | 10072 | -0.002 | -0.3495 | No | ||
| 63 | IL22 | 11948 | 10124 | -0.002 | -0.3523 | No | ||
| 64 | HECTD2 | 23876 3721 | 10346 | -0.005 | -0.3642 | No | ||
| 65 | FABP2 | 15430 | 10421 | -0.006 | -0.3681 | No | ||
| 66 | NCDN | 15756 2487 6471 | 10452 | -0.006 | -0.3697 | No | ||
| 67 | OGN | 21642 3215 | 10508 | -0.007 | -0.3726 | No | ||
| 68 | EHF | 4657 | 10541 | -0.008 | -0.3742 | No | ||
| 69 | USP2 | 19480 3052 3043 | 10584 | -0.008 | -0.3764 | No | ||
| 70 | LGI1 | 23867 | 10920 | -0.013 | -0.3944 | No | ||
| 71 | CASQ2 | 15477 | 11315 | -0.019 | -0.4155 | No | ||
| 72 | RTN1 | 4177 2099 | 11695 | -0.025 | -0.4357 | No | ||
| 73 | MSX2 | 21461 | 11709 | -0.025 | -0.4362 | No | ||
| 74 | SOX5 | 9850 1044 5480 16937 | 11803 | -0.027 | -0.4409 | No | ||
| 75 | UBXD3 | 15701 | 11966 | -0.029 | -0.4494 | No | ||
| 76 | ROS1 | 19771 | 11984 | -0.030 | -0.4500 | No | ||
| 77 | ACCN1 | 20327 1194 | 12053 | -0.031 | -0.4534 | No | ||
| 78 | SIX1 | 9821 | 12055 | -0.031 | -0.4531 | No | ||
| 79 | SATB2 | 5604 | 12242 | -0.035 | -0.4628 | No | ||
| 80 | TSHB | 15218 | 12384 | -0.038 | -0.4701 | No | ||
| 81 | ESRRG | 6447 | 12406 | -0.039 | -0.4708 | No | ||
| 82 | IL7 | 4921 | 12845 | -0.050 | -0.4940 | No | ||
| 83 | HIPK1 | 4851 | 12960 | -0.053 | -0.4996 | No | ||
| 84 | POU4F2 | 9603 5276 | 13022 | -0.055 | -0.5024 | No | ||
| 85 | BACE2 | 1738 22687 1695 | 13157 | -0.059 | -0.5090 | No | ||
| 86 | PTGFR | 15138 | 13203 | -0.061 | -0.5108 | No | ||
| 87 | HOXB8 | 9111 | 13396 | -0.068 | -0.5205 | No | ||
| 88 | CSMD3 | 10735 7938 | 13547 | -0.075 | -0.5279 | No | ||
| 89 | PLAG1 | 7148 15949 12161 | 13589 | -0.078 | -0.5293 | No | ||
| 90 | A2BP1 | 11212 1652 1689 | 13688 | -0.083 | -0.5338 | No | ||
| 91 | SORCS1 | 7184 12203 | 13764 | -0.087 | -0.5370 | No | ||
| 92 | HTR3B | 19129 3145 | 13879 | -0.093 | -0.5422 | No | ||
| 93 | MGLL | 17368 1145 | 14028 | -0.104 | -0.5491 | No | ||
| 94 | SLC26A9 | 11560 980 4067 | 14207 | -0.120 | -0.5575 | No | ||
| 95 | XYLT2 | 20288 | 14215 | -0.120 | -0.5567 | No | ||
| 96 | UBE2R2 | 16240 | 14261 | -0.125 | -0.5579 | No | ||
| 97 | HSPB2 | 19121 | 14471 | -0.149 | -0.5677 | No | ||
| 98 | TMOD4 | 1816 1884 15509 | 14542 | -0.156 | -0.5699 | No | ||
| 99 | DMD | 24295 2647 | 14621 | -0.170 | -0.5724 | No | ||
| 100 | NF1 | 5165 | 15767 | -0.530 | -0.6290 | Yes | ||
| 101 | SRPK2 | 5513 | 15782 | -0.539 | -0.6243 | Yes | ||
| 102 | SESN3 | 19561 | 15821 | -0.567 | -0.6206 | Yes | ||
| 103 | PPARGC1A | 16533 | 15891 | -0.631 | -0.6180 | Yes | ||
| 104 | FUT11 | 13084 | 15903 | -0.637 | -0.6122 | Yes | ||
| 105 | CPNE1 | 11186 | 16094 | -0.816 | -0.6142 | Yes | ||
| 106 | ZFPM1 | 18439 | 16573 | -1.314 | -0.6268 | Yes | ||
| 107 | ARHGDIB | 16950 | 16594 | -1.343 | -0.6143 | Yes | ||
| 108 | WDR23 | 22009 | 16995 | -1.897 | -0.6168 | Yes | ||
| 109 | LUC7L | 47 1520 | 17310 | -2.444 | -0.6092 | Yes | ||
| 110 | CRYAB | 19457 | 17502 | -2.871 | -0.5905 | Yes | ||
| 111 | MEF2A | 1201 5089 3492 | 17654 | -3.235 | -0.5661 | Yes | ||
| 112 | HIST1H4C | 6643 | 17664 | -3.276 | -0.5335 | Yes | ||
| 113 | SMAD5 | 21621 | 17677 | -3.317 | -0.5007 | Yes | ||
| 114 | ACTR1A | 23653 | 17730 | -3.449 | -0.4687 | Yes | ||
| 115 | TOB1 | 20703 | 17875 | -3.812 | -0.4381 | Yes | ||
| 116 | MBNL1 | 1921 15582 | 18100 | -4.631 | -0.4035 | Yes | ||
| 117 | RHOG | 17727 | 18209 | -5.063 | -0.3583 | Yes | ||
| 118 | CHD2 | 10847 | 18237 | -5.235 | -0.3069 | Yes | ||
| 119 | MBNL2 | 21932 | 18284 | -5.431 | -0.2546 | Yes | ||
| 120 | ARHGAP8 | 4268 2196 | 18306 | -5.586 | -0.1994 | Yes | ||
| 121 | RAI1 | 5355 20861 | 18390 | -6.085 | -0.1425 | Yes | ||
| 122 | HHEX | 23872 | 18391 | -6.087 | -0.0811 | Yes | ||
| 123 | LMO2 | 9281 | 18598 | -9.247 | 0.0010 | Yes |