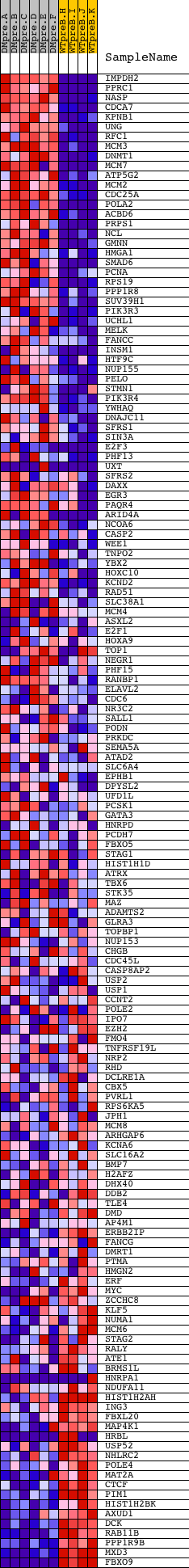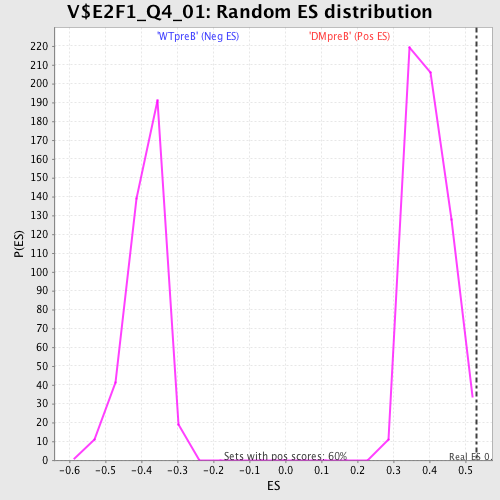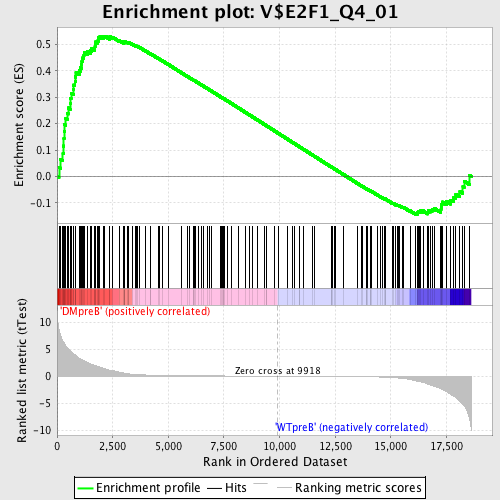
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | DMpreB |
| GeneSet | V$E2F1_Q4_01 |
| Enrichment Score (ES) | 0.53175104 |
| Normalized Enrichment Score (NES) | 1.3357314 |
| Nominal p-value | 0.006688963 |
| FDR q-value | 0.294463 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | IMPDH2 | 10730 | 85 | 8.592 | 0.0343 | Yes | ||
| 2 | PPRC1 | 23808 | 151 | 7.606 | 0.0651 | Yes | ||
| 3 | NASP | 2383 2387 6955 | 263 | 6.510 | 0.0886 | Yes | ||
| 4 | CDCA7 | 12437 | 293 | 6.274 | 0.1154 | Yes | ||
| 5 | KPNB1 | 20274 | 306 | 6.181 | 0.1427 | Yes | ||
| 6 | UNG | 10257 16744 | 315 | 6.124 | 0.1700 | Yes | ||
| 7 | RFC1 | 16527 | 335 | 5.989 | 0.1960 | Yes | ||
| 8 | MCM3 | 13991 | 390 | 5.583 | 0.2184 | Yes | ||
| 9 | DNMT1 | 19217 | 456 | 5.277 | 0.2387 | Yes | ||
| 10 | MCM7 | 9372 3568 | 497 | 5.089 | 0.2596 | Yes | ||
| 11 | ATP5G2 | 12610 | 588 | 4.682 | 0.2759 | Yes | ||
| 12 | MCM2 | 17074 | 598 | 4.627 | 0.2963 | Yes | ||
| 13 | CDC25A | 8721 | 631 | 4.494 | 0.3149 | Yes | ||
| 14 | POLA2 | 23988 | 733 | 4.174 | 0.3283 | Yes | ||
| 15 | ACBD6 | 14089 | 743 | 4.124 | 0.3465 | Yes | ||
| 16 | PRPS1 | 24233 | 804 | 3.939 | 0.3611 | Yes | ||
| 17 | NCL | 5153 13899 | 818 | 3.882 | 0.3779 | Yes | ||
| 18 | GMNN | 21513 | 848 | 3.827 | 0.3937 | Yes | ||
| 19 | HMGA1 | 23323 | 1003 | 3.313 | 0.4003 | Yes | ||
| 20 | SMAD6 | 19083 | 1038 | 3.214 | 0.4130 | Yes | ||
| 21 | PCNA | 9535 | 1100 | 3.074 | 0.4236 | Yes | ||
| 22 | RPS19 | 5398 | 1114 | 3.041 | 0.4367 | Yes | ||
| 23 | PPP1R8 | 15730 | 1138 | 2.994 | 0.4490 | Yes | ||
| 24 | SUV39H1 | 24194 | 1202 | 2.857 | 0.4585 | Yes | ||
| 25 | PIK3R3 | 5248 | 1211 | 2.839 | 0.4709 | Yes | ||
| 26 | UCHL1 | 16834 | 1369 | 2.481 | 0.4736 | Yes | ||
| 27 | MELK | 2453 16227 | 1495 | 2.254 | 0.4770 | Yes | ||
| 28 | FANCC | 4712 | 1556 | 2.184 | 0.4836 | Yes | ||
| 29 | INSM1 | 14815 | 1670 | 2.029 | 0.4867 | Yes | ||
| 30 | HTF9C | 4886 1678 | 1702 | 1.969 | 0.4939 | Yes | ||
| 31 | NUP155 | 2298 5027 | 1713 | 1.943 | 0.5022 | Yes | ||
| 32 | PELO | 8360 | 1736 | 1.906 | 0.5096 | Yes | ||
| 33 | STMN1 | 9261 | 1819 | 1.798 | 0.5133 | Yes | ||
| 34 | PIK3R4 | 19333 | 1855 | 1.740 | 0.5193 | Yes | ||
| 35 | YWHAQ | 10369 | 1863 | 1.733 | 0.5267 | Yes | ||
| 36 | DNAJC11 | 2486 6068 | 1924 | 1.635 | 0.5309 | Yes | ||
| 37 | SFRS1 | 8492 | 2072 | 1.466 | 0.5296 | Yes | ||
| 38 | SIN3A | 5442 | 2148 | 1.383 | 0.5318 | Yes | ||
| 39 | E2F3 | 21500 8874 | 2367 | 1.098 | 0.5249 | No | ||
| 40 | PHF13 | 15659 | 2372 | 1.093 | 0.5296 | No | ||
| 41 | UXT | 5837 10267 | 2499 | 0.999 | 0.5273 | No | ||
| 42 | SFRS2 | 9807 20136 | 2789 | 0.733 | 0.5150 | No | ||
| 43 | DAXX | 23290 | 2992 | 0.562 | 0.5066 | No | ||
| 44 | EGR3 | 4656 | 3013 | 0.549 | 0.5080 | No | ||
| 45 | PAQR4 | 23105 | 3014 | 0.549 | 0.5105 | No | ||
| 46 | ARID4A | 2080 6215 | 3041 | 0.533 | 0.5115 | No | ||
| 47 | NCOA6 | 2876 14382 | 3151 | 0.461 | 0.5077 | No | ||
| 48 | CASP2 | 8692 | 3192 | 0.445 | 0.5075 | No | ||
| 49 | WEE1 | 18127 | 3229 | 0.429 | 0.5075 | No | ||
| 50 | TNPO2 | 5608 | 3375 | 0.362 | 0.5013 | No | ||
| 51 | YBX2 | 20818 | 3538 | 0.316 | 0.4939 | No | ||
| 52 | HOXC10 | 22342 | 3555 | 0.312 | 0.4945 | No | ||
| 53 | KCND2 | 4941 17515 | 3623 | 0.295 | 0.4922 | No | ||
| 54 | RAD51 | 2897 14903 | 3710 | 0.271 | 0.4888 | No | ||
| 55 | SLC38A1 | 22148 | 3975 | 0.218 | 0.4754 | No | ||
| 56 | MCM4 | 22655 1708 | 4196 | 0.186 | 0.4644 | No | ||
| 57 | ASXL2 | 7957 | 4207 | 0.184 | 0.4647 | No | ||
| 58 | E2F1 | 14384 | 4565 | 0.144 | 0.4460 | No | ||
| 59 | HOXA9 | 17147 9109 1024 | 4614 | 0.139 | 0.4440 | No | ||
| 60 | TOP1 | 5790 5789 10210 | 4715 | 0.132 | 0.4392 | No | ||
| 61 | NEGR1 | 6806 11572 | 4984 | 0.113 | 0.4252 | No | ||
| 62 | PHF15 | 20467 8050 | 5571 | 0.083 | 0.3938 | No | ||
| 63 | RANBP1 | 9692 5357 | 5854 | 0.071 | 0.3789 | No | ||
| 64 | ELAVL2 | 15839 | 5942 | 0.068 | 0.3745 | No | ||
| 65 | CDC6 | 6221 6220 | 6125 | 0.063 | 0.3649 | No | ||
| 66 | NR3C2 | 18562 13798 | 6127 | 0.063 | 0.3651 | No | ||
| 67 | SALL1 | 18796 12207 | 6190 | 0.061 | 0.3620 | No | ||
| 68 | PODN | 15811 | 6202 | 0.060 | 0.3617 | No | ||
| 69 | PRKDC | 9617 22847 | 6336 | 0.057 | 0.3548 | No | ||
| 70 | SEMA5A | 22496 5423 | 6492 | 0.053 | 0.3466 | No | ||
| 71 | ATAD2 | 2268 2256 | 6586 | 0.051 | 0.3418 | No | ||
| 72 | SLC6A4 | 20770 | 6749 | 0.047 | 0.3332 | No | ||
| 73 | EPHB1 | 19024 | 6869 | 0.045 | 0.3270 | No | ||
| 74 | DPYSL2 | 3122 4561 8787 | 6945 | 0.043 | 0.3231 | No | ||
| 75 | UFD1L | 22825 | 7359 | 0.035 | 0.3009 | No | ||
| 76 | PCSK1 | 21600 | 7368 | 0.035 | 0.3007 | No | ||
| 77 | GATA3 | 9004 | 7429 | 0.034 | 0.2976 | No | ||
| 78 | HNRPD | 8645 | 7481 | 0.033 | 0.2949 | No | ||
| 79 | PCDH7 | 7029 3460 | 7483 | 0.033 | 0.2950 | No | ||
| 80 | FBXO5 | 19830 | 7525 | 0.032 | 0.2930 | No | ||
| 81 | STAG1 | 5520 9905 2987 | 7660 | 0.030 | 0.2858 | No | ||
| 82 | HIST1H1D | 4821 | 7823 | 0.027 | 0.2772 | No | ||
| 83 | ATRX | 10350 5922 2628 | 8144 | 0.023 | 0.2600 | No | ||
| 84 | TBX6 | 1993 18075 | 8478 | 0.018 | 0.2420 | No | ||
| 85 | STK35 | 14851 | 8658 | 0.016 | 0.2324 | No | ||
| 86 | MAZ | 1327 17623 | 8787 | 0.014 | 0.2255 | No | ||
| 87 | ADAMTS2 | 5706 20899 1356 | 9005 | 0.012 | 0.2138 | No | ||
| 88 | GLRA3 | 18617 | 9309 | 0.008 | 0.1974 | No | ||
| 89 | TOPBP1 | 10659 | 9405 | 0.006 | 0.1923 | No | ||
| 90 | NUP153 | 21474 | 9407 | 0.006 | 0.1923 | No | ||
| 91 | CHGB | 14835 | 9776 | 0.002 | 0.1724 | No | ||
| 92 | CDC45L | 22642 1752 | 9951 | -0.001 | 0.1630 | No | ||
| 93 | CASP8AP2 | 16253 | 10367 | -0.005 | 0.1405 | No | ||
| 94 | USP2 | 19480 3052 3043 | 10584 | -0.008 | 0.1288 | No | ||
| 95 | USP1 | 10497 10498 6051 | 10659 | -0.009 | 0.1249 | No | ||
| 96 | CCNT2 | 3993 13079 | 10873 | -0.012 | 0.1134 | No | ||
| 97 | POLE2 | 21053 | 10899 | -0.013 | 0.1121 | No | ||
| 98 | IPO7 | 6130 | 11087 | -0.015 | 0.1020 | No | ||
| 99 | EZH2 | 1092 17163 | 11493 | -0.021 | 0.0802 | No | ||
| 100 | FMO4 | 13786 | 11581 | -0.023 | 0.0756 | No | ||
| 101 | TNFRSF19L | 11498 6727 6726 | 12325 | -0.037 | 0.0355 | No | ||
| 102 | NRP2 | 5194 9486 5195 | 12343 | -0.038 | 0.0348 | No | ||
| 103 | RHD | 16042 | 12393 | -0.039 | 0.0323 | No | ||
| 104 | DCLRE1A | 23641 | 12480 | -0.041 | 0.0278 | No | ||
| 105 | CBX5 | 22108 4485 | 12505 | -0.041 | 0.0267 | No | ||
| 106 | PVRL1 | 3006 19482 12211 7192 | 12855 | -0.050 | 0.0080 | No | ||
| 107 | RPS6KA5 | 2076 21005 | 13512 | -0.073 | -0.0272 | No | ||
| 108 | JPH1 | 13994 | 13674 | -0.082 | -0.0355 | No | ||
| 109 | MCM8 | 14834 | 13711 | -0.084 | -0.0371 | No | ||
| 110 | ARHGAP6 | 24207 | 13898 | -0.095 | -0.0467 | No | ||
| 111 | KCNA6 | 664 16990 665 4938 | 13927 | -0.097 | -0.0478 | No | ||
| 112 | SLC16A2 | 24086 | 13972 | -0.100 | -0.0498 | No | ||
| 113 | BMP7 | 14324 | 14065 | -0.107 | -0.0542 | No | ||
| 114 | H2AFZ | 6957 | 14069 | -0.108 | -0.0539 | No | ||
| 115 | DHX40 | 20307 | 14135 | -0.112 | -0.0569 | No | ||
| 116 | DDB2 | 2721 2878 14525 2661 | 14421 | -0.143 | -0.0717 | No | ||
| 117 | TLE4 | 23720 3697 | 14521 | -0.154 | -0.0764 | No | ||
| 118 | DMD | 24295 2647 | 14621 | -0.170 | -0.0810 | No | ||
| 119 | AP4M1 | 3499 16656 | 14702 | -0.183 | -0.0845 | No | ||
| 120 | ERBB2IP | 3232 12234 | 14739 | -0.190 | -0.0856 | No | ||
| 121 | FANCG | 15904 | 14741 | -0.191 | -0.0848 | No | ||
| 122 | DMRT1 | 23899 | 15095 | -0.263 | -0.1027 | No | ||
| 123 | PTMA | 9657 | 15118 | -0.268 | -0.1027 | No | ||
| 124 | HMGN2 | 9095 | 15229 | -0.297 | -0.1073 | No | ||
| 125 | ERF | 4680 8914 | 15283 | -0.315 | -0.1087 | No | ||
| 126 | MYC | 22465 9435 | 15334 | -0.330 | -0.1100 | No | ||
| 127 | ZCCHC8 | 3570 7696 | 15368 | -0.339 | -0.1102 | No | ||
| 128 | KLF5 | 4456 | 15510 | -0.399 | -0.1160 | No | ||
| 129 | NUMA1 | 18167 | 15573 | -0.432 | -0.1174 | No | ||
| 130 | MCM6 | 4000 13845 4119 | 15865 | -0.608 | -0.1305 | No | ||
| 131 | STAG2 | 5521 | 16122 | -0.850 | -0.1405 | No | ||
| 132 | RALY | 2718 9690 | 16184 | -0.910 | -0.1397 | No | ||
| 133 | ATE1 | 388 17602 | 16199 | -0.926 | -0.1362 | No | ||
| 134 | BRMS1L | 21268 | 16232 | -0.966 | -0.1336 | No | ||
| 135 | HNRPA1 | 9103 4860 | 16286 | -0.999 | -0.1319 | No | ||
| 136 | NDUFA11 | 12832 | 16348 | -1.042 | -0.1305 | No | ||
| 137 | HIST1H2AH | 11414 | 16447 | -1.152 | -0.1306 | No | ||
| 138 | ING3 | 17514 1019 | 16667 | -1.449 | -0.1359 | No | ||
| 139 | FBXL20 | 13010 | 16688 | -1.488 | -0.1303 | No | ||
| 140 | MAP4K1 | 18313 | 16775 | -1.624 | -0.1276 | No | ||
| 141 | HRBL | 3452 16326 3614 | 16853 | -1.724 | -0.1240 | No | ||
| 142 | USP52 | 3397 19841 | 16964 | -1.853 | -0.1216 | No | ||
| 143 | NHLRC2 | 23828 | 17254 | -2.335 | -0.1266 | No | ||
| 144 | POLE4 | 12441 | 17259 | -2.349 | -0.1162 | No | ||
| 145 | MAT2A | 10553 10554 6099 | 17281 | -2.385 | -0.1066 | No | ||
| 146 | CTCF | 18490 | 17307 | -2.434 | -0.0969 | No | ||
| 147 | PIM1 | 23308 | 17500 | -2.869 | -0.0943 | No | ||
| 148 | HIST1H2BK | 11423 | 17694 | -3.349 | -0.0896 | No | ||
| 149 | AXUD1 | 18967 | 17830 | -3.686 | -0.0803 | No | ||
| 150 | DCK | 16808 | 17923 | -3.959 | -0.0674 | No | ||
| 151 | RAB11B | 9676 | 18106 | -4.643 | -0.0562 | No | ||
| 152 | PPP1R9B | 20698 | 18242 | -5.266 | -0.0397 | No | ||
| 153 | MXD3 | 21455 | 18295 | -5.505 | -0.0176 | No | ||
| 154 | FBXO9 | 19058 3146 | 18531 | -7.726 | 0.0046 | No |