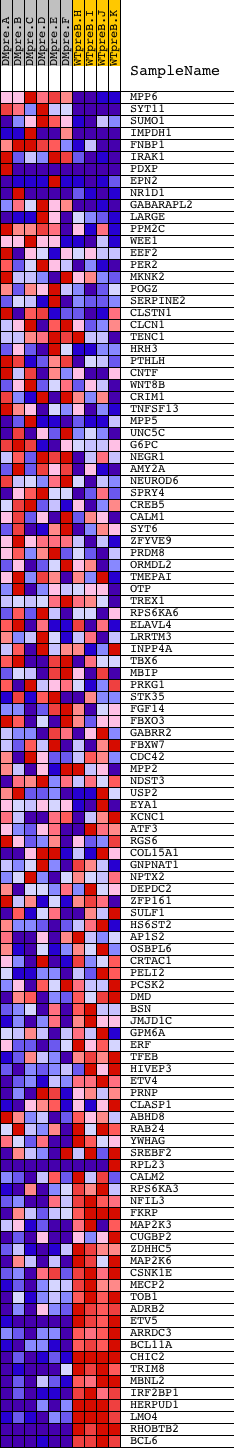
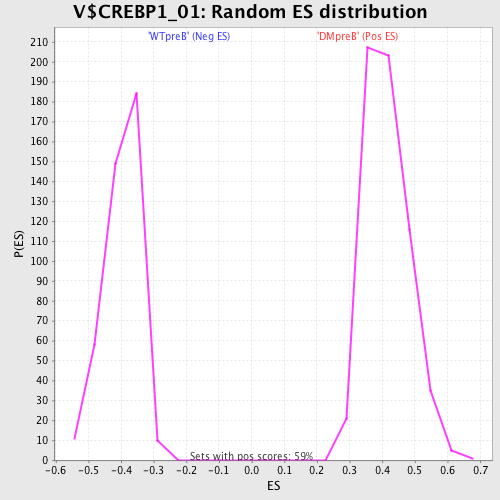
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | V$CREBP1_01 |
| Enrichment Score (ES) | -0.6310806 |
| Normalized Enrichment Score (NES) | -1.5979613 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0560399 |
| FWER p-Value | 0.311 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MPP6 | 17448 | 371 | 5.712 | 0.0275 | No | ||
| 2 | SYT11 | 15285 | 1044 | 3.199 | 0.0178 | No | ||
| 3 | SUMO1 | 5826 3943 10247 | 1636 | 2.077 | 0.0032 | No | ||
| 4 | IMPDH1 | 17197 1131 | 2099 | 1.444 | -0.0098 | No | ||
| 5 | FNBP1 | 4732 | 2202 | 1.310 | -0.0044 | No | ||
| 6 | IRAK1 | 4916 | 2334 | 1.136 | -0.0020 | No | ||
| 7 | PDXP | 22430 40 24244 | 2504 | 0.998 | -0.0028 | No | ||
| 8 | EPN2 | 20413 945 946 955 | 2611 | 0.905 | -0.0010 | No | ||
| 9 | NR1D1 | 4650 20258 949 | 2687 | 0.830 | 0.0018 | No | ||
| 10 | GABARAPL2 | 8211 13552 | 2747 | 0.769 | 0.0050 | No | ||
| 11 | LARGE | 18837 | 2749 | 0.768 | 0.0114 | No | ||
| 12 | PPM2C | 491 | 3034 | 0.536 | 0.0005 | No | ||
| 13 | WEE1 | 18127 | 3229 | 0.429 | -0.0064 | No | ||
| 14 | EEF2 | 8881 4654 8882 | 3263 | 0.412 | -0.0048 | No | ||
| 15 | PER2 | 3940 13881 | 3451 | 0.341 | -0.0121 | No | ||
| 16 | MKNK2 | 3299 9392 | 3518 | 0.322 | -0.0129 | No | ||
| 17 | POGZ | 6031 1911 | 3612 | 0.300 | -0.0155 | No | ||
| 18 | SERPINE2 | 13906 | 3920 | 0.227 | -0.0302 | No | ||
| 19 | CLSTN1 | 7241 | 4031 | 0.209 | -0.0344 | No | ||
| 20 | CLCN1 | 8744 | 4059 | 0.205 | -0.0341 | No | ||
| 21 | TENC1 | 22349 9927 | 4072 | 0.204 | -0.0331 | No | ||
| 22 | HRH3 | 14317 | 4341 | 0.168 | -0.0462 | No | ||
| 23 | PTHLH | 16931 | 4462 | 0.154 | -0.0514 | No | ||
| 24 | CNTF | 3741 8761 3680 3736 | 4532 | 0.147 | -0.0539 | No | ||
| 25 | WNT8B | 23841 | 4600 | 0.141 | -0.0563 | No | ||
| 26 | CRIM1 | 403 | 4616 | 0.139 | -0.0560 | No | ||
| 27 | TNFSF13 | 12806 | 4662 | 0.135 | -0.0573 | No | ||
| 28 | MPP5 | 21230 7078 | 4682 | 0.134 | -0.0572 | No | ||
| 29 | UNC5C | 10256 | 4877 | 0.120 | -0.0667 | No | ||
| 30 | G6PC | 20656 | 4892 | 0.119 | -0.0664 | No | ||
| 31 | NEGR1 | 6806 11572 | 4984 | 0.113 | -0.0704 | No | ||
| 32 | AMY2A | 8587 | 5243 | 0.099 | -0.0835 | No | ||
| 33 | NEUROD6 | 17138 | 5317 | 0.095 | -0.0867 | No | ||
| 34 | SPRY4 | 23448 | 5960 | 0.068 | -0.1208 | No | ||
| 35 | CREB5 | 10551 | 6052 | 0.065 | -0.1252 | No | ||
| 36 | CALM1 | 21184 | 6697 | 0.049 | -0.1596 | No | ||
| 37 | SYT6 | 13437 7042 12040 | 6823 | 0.046 | -0.1660 | No | ||
| 38 | ZFYVE9 | 10501 2449 | 6887 | 0.045 | -0.1690 | No | ||
| 39 | PRDM8 | 8084 | 7184 | 0.039 | -0.1847 | No | ||
| 40 | ORMDL2 | 19588 | 7265 | 0.037 | -0.1887 | No | ||
| 41 | TMEPAI | 51 | 7394 | 0.035 | -0.1953 | No | ||
| 42 | OTP | 21582 | 7424 | 0.034 | -0.1966 | No | ||
| 43 | TREX1 | 10219 3111 | 7470 | 0.033 | -0.1988 | No | ||
| 44 | RPS6KA6 | 12458 | 7998 | 0.025 | -0.2270 | No | ||
| 45 | ELAVL4 | 15805 4889 9137 | 8276 | 0.021 | -0.2418 | No | ||
| 46 | LRRTM3 | 19744 | 8438 | 0.019 | -0.2504 | No | ||
| 47 | INPP4A | 3985 3935 11237 4085 | 8451 | 0.018 | -0.2509 | No | ||
| 48 | TBX6 | 1993 18075 | 8478 | 0.018 | -0.2521 | No | ||
| 49 | MBIP | 21063 2072 | 8582 | 0.017 | -0.2576 | No | ||
| 50 | PRKG1 | 5289 | 8606 | 0.016 | -0.2587 | No | ||
| 51 | STK35 | 14851 | 8658 | 0.016 | -0.2613 | No | ||
| 52 | FGF14 | 21716 3005 3463 | 9073 | 0.011 | -0.2836 | No | ||
| 53 | FBXO3 | 2662 14934 | 9512 | 0.005 | -0.3072 | No | ||
| 54 | GABRR2 | 16252 | 10262 | -0.004 | -0.3476 | No | ||
| 55 | FBXW7 | 1805 11928 | 10321 | -0.005 | -0.3507 | No | ||
| 56 | CDC42 | 4503 8722 4504 2465 | 10407 | -0.006 | -0.3553 | No | ||
| 57 | MPP2 | 20209 | 10408 | -0.006 | -0.3552 | No | ||
| 58 | NDST3 | 13501 | 10544 | -0.008 | -0.3625 | No | ||
| 59 | USP2 | 19480 3052 3043 | 10584 | -0.008 | -0.3645 | No | ||
| 60 | EYA1 | 4695 4061 | 10600 | -0.009 | -0.3652 | No | ||
| 61 | KCNC1 | 9204 | 10797 | -0.011 | -0.3757 | No | ||
| 62 | ATF3 | 13720 | 10907 | -0.013 | -0.3815 | No | ||
| 63 | RGS6 | 21219 | 11206 | -0.017 | -0.3975 | No | ||
| 64 | COL15A1 | 8764 | 11314 | -0.019 | -0.4031 | No | ||
| 65 | GNPNAT1 | 12027 7031 | 11410 | -0.020 | -0.4081 | No | ||
| 66 | NPTX2 | 16630 | 11425 | -0.020 | -0.4087 | No | ||
| 67 | DEPDC2 | 14295 | 11860 | -0.027 | -0.4319 | No | ||
| 68 | ZFP161 | 5951 10381 | 11986 | -0.030 | -0.4384 | No | ||
| 69 | SULF1 | 6285 | 12809 | -0.049 | -0.4824 | No | ||
| 70 | HS6ST2 | 6939 2646 | 13091 | -0.057 | -0.4971 | No | ||
| 71 | AP1S2 | 8423 4228 | 13489 | -0.072 | -0.5180 | No | ||
| 72 | OSBPL6 | 8277 | 13549 | -0.076 | -0.5205 | No | ||
| 73 | CRTAC1 | 23673 | 14072 | -0.108 | -0.5479 | No | ||
| 74 | PELI2 | 8217 | 14133 | -0.112 | -0.5502 | No | ||
| 75 | PCSK2 | 9537 5231 14824 | 14347 | -0.135 | -0.5606 | No | ||
| 76 | DMD | 24295 2647 | 14621 | -0.170 | -0.5739 | No | ||
| 77 | BSN | 18997 | 14713 | -0.186 | -0.5773 | No | ||
| 78 | JMJD1C | 11531 19996 | 14762 | -0.195 | -0.5782 | No | ||
| 79 | GPM6A | 3834 10618 | 14806 | -0.202 | -0.5789 | No | ||
| 80 | ERF | 4680 8914 | 15283 | -0.315 | -0.6020 | No | ||
| 81 | TFEB | 23207 | 15349 | -0.334 | -0.6027 | No | ||
| 82 | HIVEP3 | 4973 | 15475 | -0.379 | -0.6063 | No | ||
| 83 | ETV4 | 20212 | 15513 | -0.403 | -0.6050 | No | ||
| 84 | PRNP | 9622 | 15543 | -0.418 | -0.6031 | No | ||
| 85 | CLASP1 | 14159 | 15606 | -0.444 | -0.6027 | No | ||
| 86 | ABHD8 | 18847 | 16132 | -0.860 | -0.6239 | Yes | ||
| 87 | RAB24 | 12584 5344 | 16181 | -0.908 | -0.6190 | Yes | ||
| 88 | YWHAG | 16339 | 16210 | -0.937 | -0.6127 | Yes | ||
| 89 | SREBF2 | 5509 | 16231 | -0.963 | -0.6057 | Yes | ||
| 90 | RPL23 | 7237 12267 20267 | 16305 | -1.005 | -0.6013 | Yes | ||
| 91 | CALM2 | 8681 | 16368 | -1.071 | -0.5957 | Yes | ||
| 92 | RPS6KA3 | 8490 | 16484 | -1.198 | -0.5920 | Yes | ||
| 93 | NFIL3 | 21463 | 16632 | -1.406 | -0.5882 | Yes | ||
| 94 | FKRP | 17953 | 16714 | -1.530 | -0.5799 | Yes | ||
| 95 | MAP2K3 | 20856 | 16736 | -1.563 | -0.5680 | Yes | ||
| 96 | CUGBP2 | 4687 8923 | 16850 | -1.721 | -0.5598 | Yes | ||
| 97 | ZDHHC5 | 14547 | 16953 | -1.839 | -0.5500 | Yes | ||
| 98 | MAP2K6 | 20614 1414 | 17087 | -2.043 | -0.5402 | Yes | ||
| 99 | CSNK1E | 6570 2211 11332 | 17205 | -2.253 | -0.5277 | Yes | ||
| 100 | MECP2 | 5088 | 17315 | -2.463 | -0.5131 | Yes | ||
| 101 | TOB1 | 20703 | 17875 | -3.812 | -0.5116 | Yes | ||
| 102 | ADRB2 | 23422 | 17922 | -3.954 | -0.4812 | Yes | ||
| 103 | ETV5 | 22630 | 17951 | -4.075 | -0.4488 | Yes | ||
| 104 | ARRDC3 | 4191 | 18030 | -4.350 | -0.4168 | Yes | ||
| 105 | BCL11A | 4691 | 18039 | -4.409 | -0.3805 | Yes | ||
| 106 | CHIC2 | 16510 | 18062 | -4.509 | -0.3441 | Yes | ||
| 107 | TRIM8 | 23824 | 18247 | -5.292 | -0.3100 | Yes | ||
| 108 | MBNL2 | 21932 | 18284 | -5.431 | -0.2668 | Yes | ||
| 109 | IRF2BP1 | 18368 | 18339 | -5.723 | -0.2221 | Yes | ||
| 110 | HERPUD1 | 18522 | 18388 | -6.080 | -0.1740 | Yes | ||
| 111 | LMO4 | 15151 | 18404 | -6.210 | -0.1232 | Yes | ||
| 112 | RHOBTB2 | 3069 6373 10902 | 18507 | -7.411 | -0.0670 | Yes | ||
| 113 | BCL6 | 22624 | 18580 | -8.747 | 0.0019 | Yes |