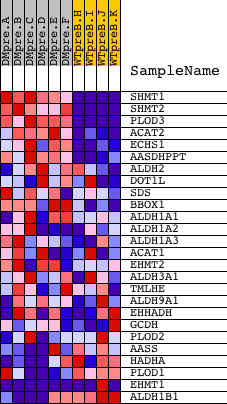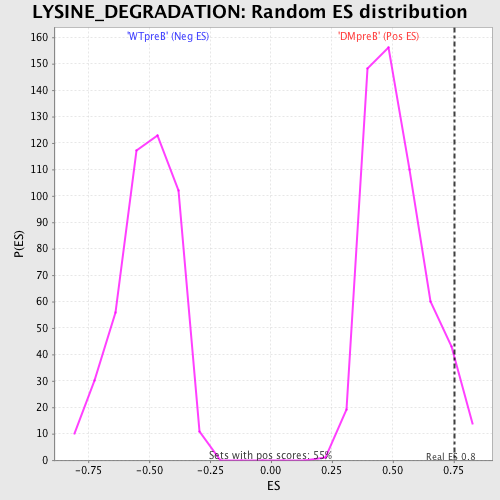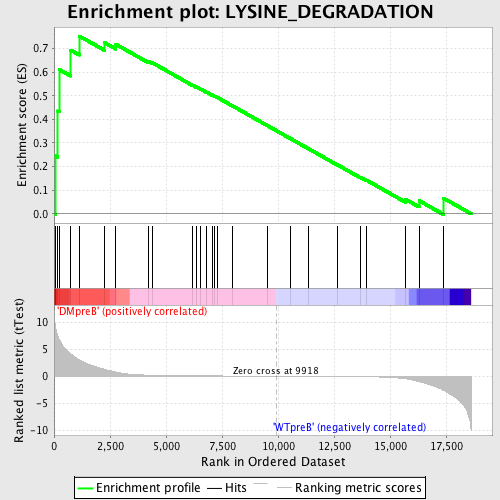
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | DMpreB |
| GeneSet | LYSINE_DEGRADATION |
| Enrichment Score (ES) | 0.75247186 |
| Normalized Enrichment Score (NES) | 1.4577919 |
| Nominal p-value | 0.049001817 |
| FDR q-value | 0.24920705 |
| FWER p-Value | 0.996 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SHMT1 | 5431 | 52 | 9.353 | 0.2461 | Yes | ||
| 2 | SHMT2 | 3307 19601 | 169 | 7.348 | 0.4354 | Yes | ||
| 3 | PLOD3 | 16666 | 229 | 6.720 | 0.6111 | Yes | ||
| 4 | ACAT2 | 8484 1537 | 742 | 4.124 | 0.6933 | Yes | ||
| 5 | ECHS1 | 17573 | 1130 | 3.007 | 0.7525 | Yes | ||
| 6 | AASDHPPT | 7467 | 2264 | 1.227 | 0.7242 | No | ||
| 7 | ALDH2 | 16384 | 2757 | 0.760 | 0.7179 | No | ||
| 8 | DOT1L | 19937 | 4192 | 0.186 | 0.6458 | No | ||
| 9 | SDS | 16722 3582 | 4373 | 0.163 | 0.6404 | No | ||
| 10 | BBOX1 | 5010 9290 | 6199 | 0.060 | 0.5439 | No | ||
| 11 | ALDH1A1 | 8569 | 6352 | 0.057 | 0.5372 | No | ||
| 12 | ALDH1A2 | 19386 | 6554 | 0.052 | 0.5278 | No | ||
| 13 | ALDH1A3 | 17802 | 6818 | 0.046 | 0.5148 | No | ||
| 14 | ACAT1 | 19114 10019 | 7054 | 0.041 | 0.5033 | No | ||
| 15 | EHMT2 | 23269 1563 | 7166 | 0.039 | 0.4984 | No | ||
| 16 | ALDH3A1 | 20854 | 7298 | 0.037 | 0.4923 | No | ||
| 17 | TMLHE | 5320 | 7978 | 0.025 | 0.4564 | No | ||
| 18 | ALDH9A1 | 14064 | 9534 | 0.005 | 0.3729 | No | ||
| 19 | EHHADH | 22635 | 10567 | -0.008 | 0.3176 | No | ||
| 20 | GCDH | 3913 18812 | 11372 | -0.020 | 0.2749 | No | ||
| 21 | PLOD2 | 19352 | 12646 | -0.045 | 0.2076 | No | ||
| 22 | AASS | 17208 1126 1026 | 13666 | -0.081 | 0.1550 | No | ||
| 23 | HADHA | 16579 | 13944 | -0.098 | 0.1427 | No | ||
| 24 | PLOD1 | 15679 | 15676 | -0.477 | 0.0623 | No | ||
| 25 | EHMT1 | 13401 14670 2756 8090 | 16292 | -1.001 | 0.0558 | No | ||
| 26 | ALDH1B1 | 16219 | 17375 | -2.598 | 0.0668 | No |