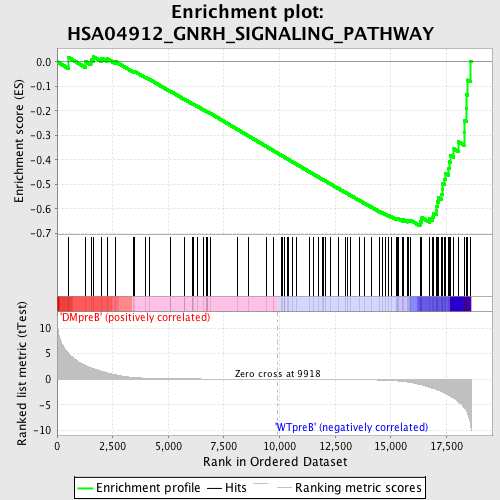
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | HSA04912_GNRH_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.66966593 |
| Normalized Enrichment Score (NES) | -1.6293658 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0327149 |
| FWER p-Value | 0.306 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PLA2G12A | 1782 7281 1839 | 517 | 4.976 | 0.0172 | No | ||
| 2 | CAMK2G | 21905 | 1270 | 2.700 | 0.0011 | No | ||
| 3 | GNAS | 9025 2963 2752 | 1524 | 2.229 | 0.0077 | No | ||
| 4 | MAPK8 | 6459 | 1644 | 2.061 | 0.0200 | No | ||
| 5 | MAPK12 | 2182 2215 22164 | 2007 | 1.545 | 0.0144 | No | ||
| 6 | CACNA1S | 14111 | 2246 | 1.244 | 0.0129 | No | ||
| 7 | HRAS | 4868 | 2610 | 0.906 | 0.0015 | No | ||
| 8 | CAMK2B | 20536 | 3423 | 0.347 | -0.0392 | No | ||
| 9 | PLA2G6 | 22209 | 3488 | 0.331 | -0.0396 | No | ||
| 10 | ADCY3 | 21329 894 | 3959 | 0.220 | -0.0630 | No | ||
| 11 | GNAQ | 4786 23909 3685 | 4136 | 0.194 | -0.0708 | No | ||
| 12 | CACNA1F | 12054 24393 | 5083 | 0.108 | -0.1208 | No | ||
| 13 | ATF4 | 22417 2239 | 5109 | 0.106 | -0.1212 | No | ||
| 14 | SRC | 5507 | 5727 | 0.076 | -0.1538 | No | ||
| 15 | CACNA1D | 4464 8675 | 5729 | 0.076 | -0.1532 | No | ||
| 16 | PRKX | 9619 5290 | 6092 | 0.064 | -0.1722 | No | ||
| 17 | PLA2G4A | 13809 | 6106 | 0.063 | -0.1723 | No | ||
| 18 | PLA2G10 | 22658 | 6131 | 0.063 | -0.1730 | No | ||
| 19 | FSHB | 14500 | 6302 | 0.058 | -0.1817 | No | ||
| 20 | MAPK13 | 23314 | 6568 | 0.052 | -0.1955 | No | ||
| 21 | CALM1 | 21184 | 6697 | 0.049 | -0.2020 | No | ||
| 22 | ADCY4 | 2980 21818 | 6720 | 0.048 | -0.2027 | No | ||
| 23 | PLCB1 | 14832 2821 | 6766 | 0.047 | -0.2047 | No | ||
| 24 | NRAS | 5191 | 6779 | 0.047 | -0.2050 | No | ||
| 25 | MAPK11 | 2264 9618 22163 | 6872 | 0.045 | -0.2095 | No | ||
| 26 | HBEGF | 23457 | 8118 | 0.023 | -0.2765 | No | ||
| 27 | MAP2K7 | 6453 | 8613 | 0.016 | -0.3030 | No | ||
| 28 | GNRHR | 4791 | 9410 | 0.006 | -0.3459 | No | ||
| 29 | PLCB4 | 9586 | 9736 | 0.002 | -0.3635 | No | ||
| 30 | LHB | 4993 | 10102 | -0.002 | -0.3831 | No | ||
| 31 | MMP2 | 18525 | 10110 | -0.002 | -0.3835 | No | ||
| 32 | PLD2 | 20803 | 10218 | -0.004 | -0.3892 | No | ||
| 33 | PLA2G1B | 9581 | 10226 | -0.004 | -0.3896 | No | ||
| 34 | CACNA1C | 1158 1166 24464 4463 8674 | 10365 | -0.005 | -0.3970 | No | ||
| 35 | CDC42 | 4503 8722 4504 2465 | 10407 | -0.006 | -0.3992 | No | ||
| 36 | PLA2G2E | 16016 | 10592 | -0.008 | -0.4090 | No | ||
| 37 | PLA2G5 | 9583 | 10744 | -0.011 | -0.4171 | No | ||
| 38 | ADCY2 | 21412 | 11356 | -0.019 | -0.4499 | No | ||
| 39 | PLA2G12B | 20016 | 11520 | -0.022 | -0.4585 | No | ||
| 40 | ADCY5 | 10312 | 11746 | -0.026 | -0.4704 | No | ||
| 41 | PLA2G2A | 16017 | 11919 | -0.028 | -0.4794 | No | ||
| 42 | PLCB2 | 5262 | 11963 | -0.029 | -0.4815 | No | ||
| 43 | ADCY8 | 22281 | 12049 | -0.031 | -0.4858 | No | ||
| 44 | PRKACA | 18549 3844 | 12270 | -0.036 | -0.4973 | No | ||
| 45 | PLA2G3 | 20968 | 12669 | -0.046 | -0.5184 | No | ||
| 46 | PLD1 | 15624 | 12945 | -0.053 | -0.5328 | No | ||
| 47 | SOS1 | 5476 | 13046 | -0.056 | -0.5377 | No | ||
| 48 | ADCY6 | 22139 2283 8551 | 13201 | -0.061 | -0.5454 | No | ||
| 49 | MAPK10 | 11169 | 13581 | -0.077 | -0.5652 | No | ||
| 50 | MAP3K4 | 23126 | 13836 | -0.091 | -0.5780 | No | ||
| 51 | GNRH1 | 125 | 14109 | -0.110 | -0.5917 | No | ||
| 52 | CAMK2A | 2024 23541 1980 | 14506 | -0.152 | -0.6117 | No | ||
| 53 | PLCB3 | 23799 | 14608 | -0.167 | -0.6157 | No | ||
| 54 | PLA2G2D | 16018 | 14754 | -0.193 | -0.6217 | No | ||
| 55 | MAP2K2 | 19933 | 14908 | -0.221 | -0.6280 | No | ||
| 56 | EGFR | 1329 20944 | 15017 | -0.243 | -0.6316 | No | ||
| 57 | MAPK7 | 1381 20414 | 15238 | -0.299 | -0.6408 | No | ||
| 58 | MAP2K1 | 19082 | 15318 | -0.325 | -0.6421 | No | ||
| 59 | PLA2G2F | 15700 | 15352 | -0.335 | -0.6408 | No | ||
| 60 | MAP3K2 | 11165 | 15532 | -0.412 | -0.6468 | No | ||
| 61 | CALML3 | 21553 | 15551 | -0.422 | -0.6439 | No | ||
| 62 | ITPR2 | 9194 | 15731 | -0.509 | -0.6490 | No | ||
| 63 | CGA | 16246 | 15780 | -0.538 | -0.6467 | No | ||
| 64 | MMP14 | 22020 | 15895 | -0.631 | -0.6471 | No | ||
| 65 | MAPK9 | 1233 20903 1383 | 16314 | -1.011 | -0.6605 | Yes | ||
| 66 | ADCY9 | 22680 | 16343 | -1.039 | -0.6526 | Yes | ||
| 67 | CALM2 | 8681 | 16368 | -1.071 | -0.6442 | Yes | ||
| 68 | ADCY7 | 8552 448 | 16398 | -1.096 | -0.6358 | Yes | ||
| 69 | MAP2K3 | 20856 | 16736 | -1.563 | -0.6398 | Yes | ||
| 70 | ITPR3 | 9195 | 16893 | -1.758 | -0.6323 | Yes | ||
| 71 | MAPK3 | 6458 11170 | 16931 | -1.809 | -0.6179 | Yes | ||
| 72 | MAP2K4 | 20405 | 17051 | -1.981 | -0.6063 | Yes | ||
| 73 | CALM3 | 8682 | 17069 | -2.011 | -0.5890 | Yes | ||
| 74 | MAP2K6 | 20614 1414 | 17087 | -2.043 | -0.5714 | Yes | ||
| 75 | RAF1 | 17035 | 17131 | -2.125 | -0.5544 | Yes | ||
| 76 | PRKCD | 21897 | 17290 | -2.395 | -0.5412 | Yes | ||
| 77 | SOS2 | 21049 | 17328 | -2.499 | -0.5206 | Yes | ||
| 78 | PRKCA | 20174 | 17337 | -2.525 | -0.4981 | Yes | ||
| 79 | MAPK14 | 23313 | 17421 | -2.682 | -0.4783 | Yes | ||
| 80 | ITPR1 | 17341 | 17446 | -2.732 | -0.4548 | Yes | ||
| 81 | GRB2 | 20149 | 17590 | -3.055 | -0.4348 | Yes | ||
| 82 | PRKACB | 15140 | 17626 | -3.163 | -0.4080 | Yes | ||
| 83 | JUN | 15832 | 17687 | -3.335 | -0.3810 | Yes | ||
| 84 | GNA11 | 3300 3345 19670 | 17835 | -3.697 | -0.3554 | Yes | ||
| 85 | MAP3K3 | 20626 | 18041 | -4.417 | -0.3264 | Yes | ||
| 86 | PRKCB1 | 1693 9574 | 18292 | -5.486 | -0.2901 | Yes | ||
| 87 | PTK2B | 21776 | 18298 | -5.513 | -0.2404 | Yes | ||
| 88 | KRAS | 9247 | 18396 | -6.150 | -0.1898 | Yes | ||
| 89 | MAP3K1 | 21348 | 18411 | -6.282 | -0.1336 | Yes | ||
| 90 | CAMK2D | 4232 | 18463 | -6.834 | -0.0744 | Yes | ||
| 91 | MAPK1 | 1642 11167 | 18592 | -9.104 | 0.0013 | Yes |

