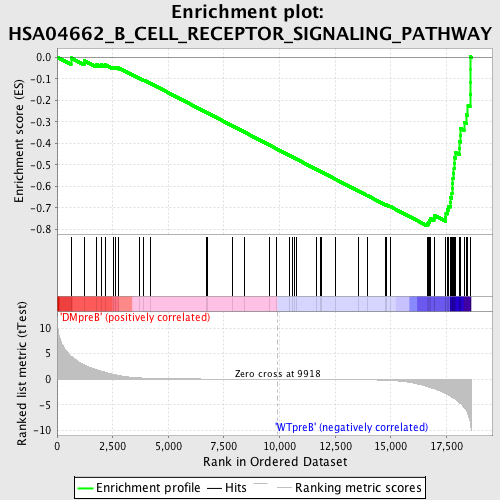
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | HSA04662_B_CELL_RECEPTOR_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.7844078 |
| Normalized Enrichment Score (NES) | -1.8094206 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0026699998 |
| FWER p-Value | 0.0020 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PIK3CB | 19030 | 626 | 4.520 | -0.0035 | No | ||
| 2 | PIK3R3 | 5248 | 1211 | 2.839 | -0.0159 | No | ||
| 3 | BLNK | 23681 3691 | 1758 | 1.877 | -0.0328 | No | ||
| 4 | MALT1 | 6274 | 1985 | 1.573 | -0.0344 | No | ||
| 5 | CHUK | 23665 | 2157 | 1.372 | -0.0344 | No | ||
| 6 | PPP3CA | 1863 5284 | 2520 | 0.993 | -0.0473 | No | ||
| 7 | HRAS | 4868 | 2610 | 0.906 | -0.0460 | No | ||
| 8 | IKBKB | 4907 | 2775 | 0.746 | -0.0499 | No | ||
| 9 | VAV3 | 1774 1848 15443 | 3695 | 0.275 | -0.0976 | No | ||
| 10 | PIK3R2 | 18850 | 3888 | 0.232 | -0.1064 | No | ||
| 11 | AKT1 | 8568 | 4218 | 0.183 | -0.1229 | No | ||
| 12 | AKT2 | 4365 4366 | 6728 | 0.048 | -0.2578 | No | ||
| 13 | NRAS | 5191 | 6779 | 0.047 | -0.2601 | No | ||
| 14 | IFITM1 | 18017 7578 12724 | 7893 | 0.026 | -0.3199 | No | ||
| 15 | PIK3R5 | 11507 | 8444 | 0.019 | -0.3495 | No | ||
| 16 | NFKB2 | 23810 | 9535 | 0.005 | -0.4082 | No | ||
| 17 | PPP3R2 | 9612 | 9858 | 0.001 | -0.4255 | No | ||
| 18 | LYN | 16281 | 10463 | -0.007 | -0.4580 | No | ||
| 19 | CR2 | 3942 13707 | 10585 | -0.008 | -0.4645 | No | ||
| 20 | RAC3 | 20561 | 10656 | -0.009 | -0.4682 | No | ||
| 21 | NFATC4 | 22002 | 10757 | -0.011 | -0.4735 | No | ||
| 22 | PPP3R1 | 9611 489 | 11661 | -0.024 | -0.5220 | No | ||
| 23 | NFATC2 | 5168 2866 | 11816 | -0.027 | -0.5301 | No | ||
| 24 | IKBKG | 2570 2562 4908 | 11885 | -0.028 | -0.5336 | No | ||
| 25 | PIK3R1 | 3170 | 12502 | -0.041 | -0.5665 | No | ||
| 26 | PIK3CA | 9562 | 13540 | -0.075 | -0.6219 | No | ||
| 27 | NFAT5 | 3921 7037 12036 | 13961 | -0.099 | -0.6439 | No | ||
| 28 | NFKBIB | 17906 | 14778 | -0.198 | -0.6866 | No | ||
| 29 | FOS | 21202 | 14800 | -0.201 | -0.6863 | No | ||
| 30 | VAV2 | 5848 2670 | 14978 | -0.236 | -0.6943 | No | ||
| 31 | RAC1 | 16302 | 16651 | -1.434 | -0.7748 | Yes | ||
| 32 | RASGRP3 | 10767 | 16689 | -1.489 | -0.7668 | Yes | ||
| 33 | PIK3CD | 9563 | 16730 | -1.548 | -0.7586 | Yes | ||
| 34 | AKT3 | 13739 982 | 16769 | -1.612 | -0.7499 | Yes | ||
| 35 | PPP3CB | 5285 | 16941 | -1.823 | -0.7469 | Yes | ||
| 36 | PPP3CC | 21763 | 16961 | -1.850 | -0.7355 | Yes | ||
| 37 | NFATC1 | 23398 1999 5167 9455 1985 1957 | 17449 | -2.738 | -0.7434 | Yes | ||
| 38 | CARD11 | 8436 | 17478 | -2.819 | -0.7260 | Yes | ||
| 39 | CD79B | 20185 1309 | 17530 | -2.928 | -0.7092 | Yes | ||
| 40 | GSK3B | 22761 | 17596 | -3.078 | -0.6921 | Yes | ||
| 41 | CD19 | 17640 | 17676 | -3.314 | -0.6741 | Yes | ||
| 42 | JUN | 15832 | 17687 | -3.335 | -0.6523 | Yes | ||
| 43 | PLCG2 | 18453 | 17743 | -3.480 | -0.6320 | Yes | ||
| 44 | CD22 | 17881 | 17755 | -3.507 | -0.6091 | Yes | ||
| 45 | NFATC3 | 5169 | 17772 | -3.544 | -0.5862 | Yes | ||
| 46 | NFKB1 | 15160 | 17789 | -3.591 | -0.5630 | Yes | ||
| 47 | CD79A | 18342 | 17804 | -3.629 | -0.5394 | Yes | ||
| 48 | NFKBIA | 21065 | 17838 | -3.699 | -0.5164 | Yes | ||
| 49 | BTK | 24061 | 17851 | -3.733 | -0.4921 | Yes | ||
| 50 | CD81 | 8719 | 17871 | -3.794 | -0.4677 | Yes | ||
| 51 | INPP5D | 14198 | 17891 | -3.853 | -0.4429 | Yes | ||
| 52 | SYK | 21636 | 18074 | -4.548 | -0.4223 | Yes | ||
| 53 | BCL10 | 15397 | 18081 | -4.569 | -0.3920 | Yes | ||
| 54 | FCGR2B | 8959 | 18125 | -4.712 | -0.3627 | Yes | ||
| 55 | VAV1 | 23173 | 18136 | -4.751 | -0.3314 | Yes | ||
| 56 | PRKCB1 | 1693 9574 | 18292 | -5.486 | -0.3030 | Yes | ||
| 57 | KRAS | 9247 | 18396 | -6.150 | -0.2674 | Yes | ||
| 58 | RAC2 | 22217 | 18467 | -6.932 | -0.2247 | Yes | ||
| 59 | PTPN6 | 17002 | 18561 | -8.216 | -0.1747 | Yes | ||
| 60 | NFKBIE | 23225 1556 | 18579 | -8.741 | -0.1171 | Yes | ||
| 61 | PIK3CG | 6635 | 18583 | -8.846 | -0.0580 | Yes | ||
| 62 | CD72 | 8718 | 18585 | -8.914 | 0.0017 | Yes |

