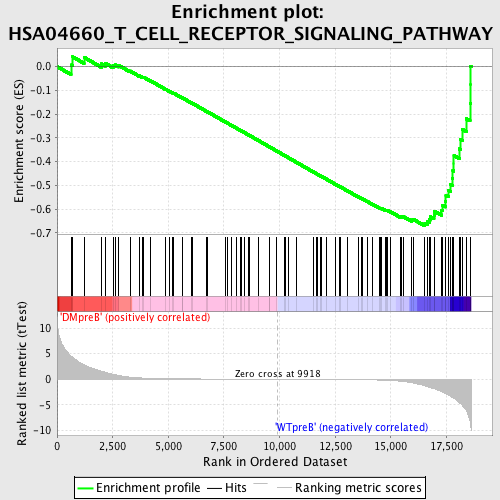
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | HSA04660_T_CELL_RECEPTOR_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.67080677 |
| Normalized Enrichment Score (NES) | -1.6474696 |
| Nominal p-value | 0.0026525198 |
| FDR q-value | 0.030807048 |
| FWER p-Value | 0.224 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PIK3CB | 19030 | 626 | 4.520 | 0.0067 | No | ||
| 2 | PDK1 | 14987 | 702 | 4.264 | 0.0409 | No | ||
| 3 | PIK3R3 | 5248 | 1211 | 2.839 | 0.0389 | No | ||
| 4 | MALT1 | 6274 | 1985 | 1.573 | 0.0112 | No | ||
| 5 | CHUK | 23665 | 2157 | 1.372 | 0.0143 | No | ||
| 6 | PPP3CA | 1863 5284 | 2520 | 0.993 | 0.0037 | No | ||
| 7 | HRAS | 4868 | 2610 | 0.906 | 0.0070 | No | ||
| 8 | IKBKB | 4907 | 2775 | 0.746 | 0.0048 | No | ||
| 9 | CDK4 | 3424 19859 | 3277 | 0.401 | -0.0186 | No | ||
| 10 | VAV3 | 1774 1848 15443 | 3695 | 0.275 | -0.0387 | No | ||
| 11 | CD3D | 19473 | 3849 | 0.241 | -0.0448 | No | ||
| 12 | PIK3R2 | 18850 | 3888 | 0.232 | -0.0448 | No | ||
| 13 | AKT1 | 8568 | 4218 | 0.183 | -0.0609 | No | ||
| 14 | IL4 | 9174 | 4886 | 0.119 | -0.0958 | No | ||
| 15 | FYN | 3375 3395 20052 | 5072 | 0.109 | -0.1048 | No | ||
| 16 | ITK | 4934 | 5197 | 0.102 | -0.1106 | No | ||
| 17 | CD4 | 16999 | 5252 | 0.099 | -0.1126 | No | ||
| 18 | PAK3 | 9528 | 5621 | 0.081 | -0.1318 | No | ||
| 19 | RASGRP1 | 5360 14476 9694 | 5625 | 0.080 | -0.1312 | No | ||
| 20 | IL5 | 20884 10220 | 6018 | 0.066 | -0.1518 | No | ||
| 21 | GRAP2 | 5113 9398 | 6102 | 0.063 | -0.1557 | No | ||
| 22 | AKT2 | 4365 4366 | 6728 | 0.048 | -0.1890 | No | ||
| 23 | NRAS | 5191 | 6779 | 0.047 | -0.1913 | No | ||
| 24 | PAK1 | 9527 | 7590 | 0.031 | -0.2347 | No | ||
| 25 | IL10 | 14145 1510 1553 22902 | 7653 | 0.030 | -0.2378 | No | ||
| 26 | NCK1 | 9447 5152 | 7819 | 0.028 | -0.2465 | No | ||
| 27 | CD3E | 8714 | 8077 | 0.024 | -0.2601 | No | ||
| 28 | RHOA | 8624 4409 4410 | 8261 | 0.021 | -0.2698 | No | ||
| 29 | CD40LG | 24330 | 8297 | 0.020 | -0.2715 | No | ||
| 30 | PIK3R5 | 11507 | 8444 | 0.019 | -0.2792 | No | ||
| 31 | IL2 | 15354 | 8580 | 0.017 | -0.2864 | No | ||
| 32 | CD28 | 14239 4092 | 8632 | 0.016 | -0.2890 | No | ||
| 33 | CD247 | 4498 8715 | 9036 | 0.011 | -0.3106 | No | ||
| 34 | NFKB2 | 23810 | 9535 | 0.005 | -0.3375 | No | ||
| 35 | PAK7 | 14417 | 9553 | 0.005 | -0.3383 | No | ||
| 36 | PPP3R2 | 9612 | 9858 | 0.001 | -0.3547 | No | ||
| 37 | ICOS | 12013 | 10231 | -0.004 | -0.3748 | No | ||
| 38 | CBLC | 17938 | 10261 | -0.004 | -0.3763 | No | ||
| 39 | CDC42 | 4503 8722 4504 2465 | 10407 | -0.006 | -0.3841 | No | ||
| 40 | NFATC4 | 22002 | 10757 | -0.011 | -0.4028 | No | ||
| 41 | CD3G | 19139 | 11522 | -0.022 | -0.4439 | No | ||
| 42 | PPP3R1 | 9611 489 | 11661 | -0.024 | -0.4511 | No | ||
| 43 | CTLA4 | 14238 | 11696 | -0.025 | -0.4527 | No | ||
| 44 | NFATC2 | 5168 2866 | 11816 | -0.027 | -0.4589 | No | ||
| 45 | IKBKG | 2570 2562 4908 | 11885 | -0.028 | -0.4623 | No | ||
| 46 | PDCD1 | 13870 | 12086 | -0.032 | -0.4728 | No | ||
| 47 | PIK3R1 | 3170 | 12502 | -0.041 | -0.4949 | No | ||
| 48 | PLCG1 | 14753 | 12675 | -0.046 | -0.5037 | No | ||
| 49 | CSF2 | 20454 | 12732 | -0.047 | -0.5063 | No | ||
| 50 | SOS1 | 5476 | 13046 | -0.056 | -0.5227 | No | ||
| 51 | PIK3CA | 9562 | 13540 | -0.075 | -0.5487 | No | ||
| 52 | TNF | 23004 | 13668 | -0.082 | -0.5548 | No | ||
| 53 | CD8B | 4502 17418 | 13714 | -0.084 | -0.5565 | No | ||
| 54 | NFAT5 | 3921 7037 12036 | 13961 | -0.099 | -0.5689 | No | ||
| 55 | LAT | 17643 | 14176 | -0.117 | -0.5794 | No | ||
| 56 | LCP2 | 4988 9268 | 14508 | -0.152 | -0.5959 | No | ||
| 57 | ZAP70 | 14271 4042 | 14538 | -0.156 | -0.5960 | No | ||
| 58 | CBL | 19154 | 14602 | -0.166 | -0.5979 | No | ||
| 59 | NFKBIB | 17906 | 14778 | -0.198 | -0.6056 | No | ||
| 60 | FOS | 21202 | 14800 | -0.201 | -0.6049 | No | ||
| 61 | PRKCQ | 2873 2831 | 14829 | -0.205 | -0.6046 | No | ||
| 62 | VAV2 | 5848 2670 | 14978 | -0.236 | -0.6105 | No | ||
| 63 | LCK | 15746 | 15440 | -0.368 | -0.6321 | No | ||
| 64 | IFNG | 19869 | 15478 | -0.380 | -0.6307 | No | ||
| 65 | PTPRC | 5327 9662 | 15564 | -0.428 | -0.6314 | No | ||
| 66 | PAK2 | 10310 5875 | 15938 | -0.671 | -0.6456 | No | ||
| 67 | MAP3K8 | 23495 | 16016 | -0.736 | -0.6431 | No | ||
| 68 | NCK2 | 9448 | 16530 | -1.250 | -0.6596 | Yes | ||
| 69 | PAK4 | 17909 | 16640 | -1.423 | -0.6527 | Yes | ||
| 70 | PIK3CD | 9563 | 16730 | -1.548 | -0.6437 | Yes | ||
| 71 | AKT3 | 13739 982 | 16769 | -1.612 | -0.6313 | Yes | ||
| 72 | PPP3CB | 5285 | 16941 | -1.823 | -0.6242 | Yes | ||
| 73 | PPP3CC | 21763 | 16961 | -1.850 | -0.6086 | Yes | ||
| 74 | TEC | 16514 | 17287 | -2.394 | -0.6047 | Yes | ||
| 75 | SOS2 | 21049 | 17328 | -2.499 | -0.5845 | Yes | ||
| 76 | NFATC1 | 23398 1999 5167 9455 1985 1957 | 17449 | -2.738 | -0.5664 | Yes | ||
| 77 | CARD11 | 8436 | 17478 | -2.819 | -0.5427 | Yes | ||
| 78 | GRB2 | 20149 | 17590 | -3.055 | -0.5213 | Yes | ||
| 79 | JUN | 15832 | 17687 | -3.335 | -0.4966 | Yes | ||
| 80 | NFATC3 | 5169 | 17772 | -3.544 | -0.4694 | Yes | ||
| 81 | NFKB1 | 15160 | 17789 | -3.591 | -0.4381 | Yes | ||
| 82 | MAP3K14 | 11998 | 17837 | -3.699 | -0.4075 | Yes | ||
| 83 | NFKBIA | 21065 | 17838 | -3.699 | -0.3743 | Yes | ||
| 84 | BCL10 | 15397 | 18081 | -4.569 | -0.3465 | Yes | ||
| 85 | VAV1 | 23173 | 18136 | -4.751 | -0.3068 | Yes | ||
| 86 | CBLB | 5531 22734 | 18225 | -5.154 | -0.2654 | Yes | ||
| 87 | KRAS | 9247 | 18396 | -6.150 | -0.2195 | Yes | ||
| 88 | PTPN6 | 17002 | 18561 | -8.216 | -0.1547 | Yes | ||
| 89 | NFKBIE | 23225 1556 | 18579 | -8.741 | -0.0773 | Yes | ||
| 90 | PIK3CG | 6635 | 18583 | -8.846 | 0.0018 | Yes |

