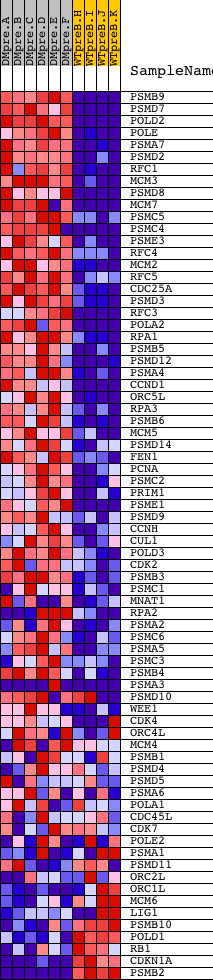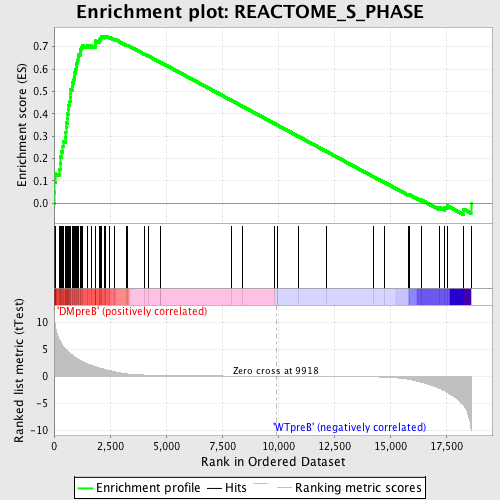
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | DMpreB |
| GeneSet | REACTOME_S_PHASE |
| Enrichment Score (ES) | 0.7472578 |
| Normalized Enrichment Score (NES) | 1.7206053 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0019624557 |
| FWER p-Value | 0.021 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSMB9 | 23021 | 15 | 10.811 | 0.0482 | Yes | ||
| 2 | PSMD7 | 18752 3825 | 28 | 10.182 | 0.0936 | Yes | ||
| 3 | POLD2 | 20537 | 74 | 8.897 | 0.1315 | Yes | ||
| 4 | POLE | 16755 | 244 | 6.625 | 0.1524 | Yes | ||
| 5 | PSMA7 | 14318 | 278 | 6.350 | 0.1794 | Yes | ||
| 6 | PSMD2 | 10137 5724 | 288 | 6.286 | 0.2073 | Yes | ||
| 7 | RFC1 | 16527 | 335 | 5.989 | 0.2320 | Yes | ||
| 8 | MCM3 | 13991 | 390 | 5.583 | 0.2544 | Yes | ||
| 9 | PSMD8 | 7166 | 418 | 5.413 | 0.2774 | Yes | ||
| 10 | MCM7 | 9372 3568 | 497 | 5.089 | 0.2963 | Yes | ||
| 11 | PSMC5 | 1348 20624 | 522 | 4.954 | 0.3174 | Yes | ||
| 12 | PSMC4 | 2141 17915 | 531 | 4.907 | 0.3392 | Yes | ||
| 13 | PSME3 | 20657 | 553 | 4.832 | 0.3599 | Yes | ||
| 14 | RFC4 | 1735 22627 | 589 | 4.674 | 0.3792 | Yes | ||
| 15 | MCM2 | 17074 | 598 | 4.627 | 0.3997 | Yes | ||
| 16 | RFC5 | 13005 7791 | 625 | 4.526 | 0.4188 | Yes | ||
| 17 | CDC25A | 8721 | 631 | 4.494 | 0.4389 | Yes | ||
| 18 | PSMD3 | 5803 | 692 | 4.290 | 0.4551 | Yes | ||
| 19 | RFC3 | 12786 | 722 | 4.193 | 0.4725 | Yes | ||
| 20 | POLA2 | 23988 | 733 | 4.174 | 0.4909 | Yes | ||
| 21 | RPA1 | 20349 | 739 | 4.140 | 0.5094 | Yes | ||
| 22 | PSMB5 | 9633 | 809 | 3.917 | 0.5234 | Yes | ||
| 23 | PSMD12 | 20621 | 821 | 3.878 | 0.5404 | Yes | ||
| 24 | PSMA4 | 11179 | 882 | 3.710 | 0.5539 | Yes | ||
| 25 | CCND1 | 4487 4488 8707 17535 | 915 | 3.583 | 0.5684 | Yes | ||
| 26 | ORC5L | 11173 3595 | 924 | 3.572 | 0.5842 | Yes | ||
| 27 | RPA3 | 12667 | 963 | 3.441 | 0.5977 | Yes | ||
| 28 | PSMB6 | 9634 | 981 | 3.372 | 0.6121 | Yes | ||
| 29 | MCM5 | 18564 | 1005 | 3.303 | 0.6258 | Yes | ||
| 30 | PSMD14 | 15005 | 1037 | 3.216 | 0.6387 | Yes | ||
| 31 | FEN1 | 8961 | 1068 | 3.138 | 0.6513 | Yes | ||
| 32 | PCNA | 9535 | 1100 | 3.074 | 0.6635 | Yes | ||
| 33 | PSMC2 | 16909 | 1164 | 2.940 | 0.6735 | Yes | ||
| 34 | PRIM1 | 19847 | 1169 | 2.930 | 0.6865 | Yes | ||
| 35 | PSME1 | 5300 9637 | 1201 | 2.861 | 0.6978 | Yes | ||
| 36 | PSMD9 | 3461 7393 | 1275 | 2.691 | 0.7060 | Yes | ||
| 37 | CCNH | 7322 | 1482 | 2.274 | 0.7052 | Yes | ||
| 38 | CUL1 | 17462 | 1686 | 2.000 | 0.7033 | Yes | ||
| 39 | POLD3 | 17742 | 1845 | 1.754 | 0.7028 | Yes | ||
| 40 | CDK2 | 3438 3373 19592 3322 | 1846 | 1.753 | 0.7107 | Yes | ||
| 41 | PSMB3 | 11180 | 1849 | 1.748 | 0.7185 | Yes | ||
| 42 | PSMC1 | 9635 | 1857 | 1.738 | 0.7260 | Yes | ||
| 43 | MNAT1 | 9396 2161 | 2016 | 1.537 | 0.7244 | Yes | ||
| 44 | RPA2 | 2330 16057 | 2021 | 1.531 | 0.7312 | Yes | ||
| 45 | PSMA2 | 9631 | 2075 | 1.465 | 0.7349 | Yes | ||
| 46 | PSMC6 | 7379 | 2094 | 1.448 | 0.7405 | Yes | ||
| 47 | PSMA5 | 6464 | 2098 | 1.446 | 0.7469 | Yes | ||
| 48 | PSMC3 | 9636 | 2250 | 1.240 | 0.7444 | Yes | ||
| 49 | PSMB4 | 15252 | 2297 | 1.181 | 0.7473 | Yes | ||
| 50 | PSMA3 | 9632 5298 | 2493 | 1.000 | 0.7413 | No | ||
| 51 | PSMD10 | 2578 2597 24046 | 2709 | 0.811 | 0.7333 | No | ||
| 52 | WEE1 | 18127 | 3229 | 0.429 | 0.7073 | No | ||
| 53 | CDK4 | 3424 19859 | 3277 | 0.401 | 0.7066 | No | ||
| 54 | ORC4L | 11172 6460 | 4035 | 0.208 | 0.6667 | No | ||
| 55 | MCM4 | 22655 1708 | 4196 | 0.186 | 0.6589 | No | ||
| 56 | PSMB1 | 23118 | 4750 | 0.128 | 0.6297 | No | ||
| 57 | PSMD4 | 15251 | 4763 | 0.127 | 0.6296 | No | ||
| 58 | PSMD5 | 2774 14612 | 7934 | 0.026 | 0.4588 | No | ||
| 59 | PSMA6 | 21270 | 8391 | 0.019 | 0.4343 | No | ||
| 60 | POLA1 | 24112 | 9841 | 0.001 | 0.3561 | No | ||
| 61 | CDC45L | 22642 1752 | 9951 | -0.001 | 0.3503 | No | ||
| 62 | CDK7 | 21365 | 9956 | -0.001 | 0.3500 | No | ||
| 63 | POLE2 | 21053 | 10899 | -0.013 | 0.2993 | No | ||
| 64 | PSMA1 | 1627 17669 | 12144 | -0.033 | 0.2324 | No | ||
| 65 | PSMD11 | 12772 7600 | 14271 | -0.126 | 0.1183 | No | ||
| 66 | ORC2L | 385 13949 | 14740 | -0.190 | 0.0939 | No | ||
| 67 | ORC1L | 327 16144 | 15837 | -0.579 | 0.0374 | No | ||
| 68 | MCM6 | 4000 13845 4119 | 15865 | -0.608 | 0.0387 | No | ||
| 69 | LIG1 | 18388 1749 1493 | 16393 | -1.092 | 0.0152 | No | ||
| 70 | PSMB10 | 5299 18761 | 17214 | -2.264 | -0.0187 | No | ||
| 71 | POLD1 | 17847 | 17439 | -2.709 | -0.0185 | No | ||
| 72 | RB1 | 21754 | 17554 | -2.982 | -0.0112 | No | ||
| 73 | CDKN1A | 4511 8729 | 18269 | -5.369 | -0.0254 | No | ||
| 74 | PSMB2 | 2324 16078 | 18611 | -9.723 | 0.0003 | No |