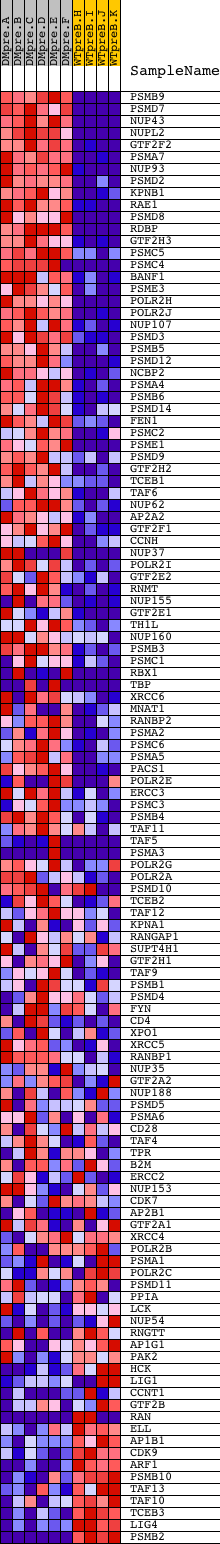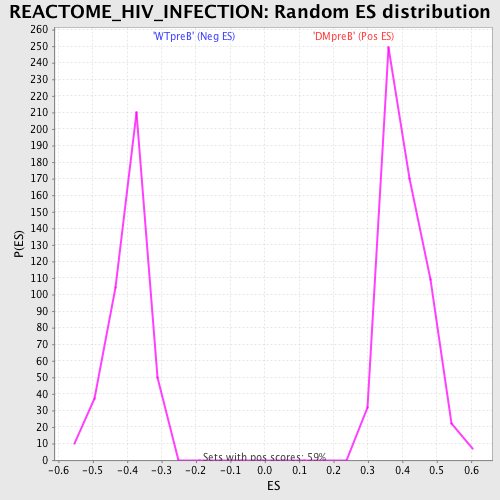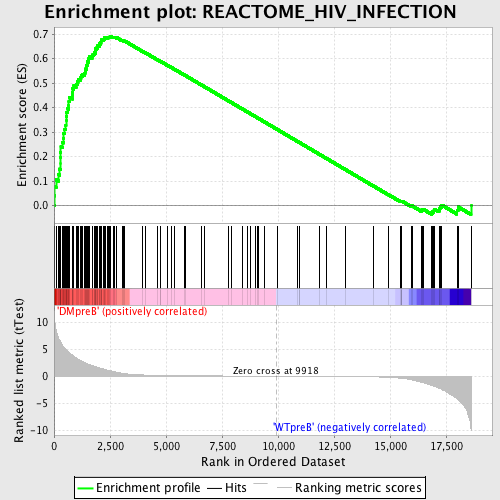
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | DMpreB |
| GeneSet | REACTOME_HIV_INFECTION |
| Enrichment Score (ES) | 0.6917594 |
| Normalized Enrichment Score (NES) | 1.7034897 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.003213642 |
| FWER p-Value | 0.041 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSMB9 | 23021 | 15 | 10.811 | 0.0397 | Yes | ||
| 2 | PSMD7 | 18752 3825 | 28 | 10.182 | 0.0773 | Yes | ||
| 3 | NUP43 | 20094 | 90 | 8.530 | 0.1060 | Yes | ||
| 4 | NUPL2 | 6072 | 203 | 6.986 | 0.1261 | Yes | ||
| 5 | GTF2F2 | 21750 | 236 | 6.685 | 0.1495 | Yes | ||
| 6 | PSMA7 | 14318 | 278 | 6.350 | 0.1711 | Yes | ||
| 7 | NUP93 | 7762 | 280 | 6.335 | 0.1948 | Yes | ||
| 8 | PSMD2 | 10137 5724 | 288 | 6.286 | 0.2180 | Yes | ||
| 9 | KPNB1 | 20274 | 306 | 6.181 | 0.2403 | Yes | ||
| 10 | RAE1 | 12395 | 381 | 5.644 | 0.2574 | Yes | ||
| 11 | PSMD8 | 7166 | 418 | 5.413 | 0.2758 | Yes | ||
| 12 | RDBP | 6589 | 428 | 5.370 | 0.2954 | Yes | ||
| 13 | GTF2H3 | 5551 | 477 | 5.199 | 0.3124 | Yes | ||
| 14 | PSMC5 | 1348 20624 | 522 | 4.954 | 0.3286 | Yes | ||
| 15 | PSMC4 | 2141 17915 | 531 | 4.907 | 0.3465 | Yes | ||
| 16 | BANF1 | 6216 10706 3760 | 537 | 4.890 | 0.3646 | Yes | ||
| 17 | PSME3 | 20657 | 553 | 4.832 | 0.3819 | Yes | ||
| 18 | POLR2H | 10888 | 611 | 4.591 | 0.3961 | Yes | ||
| 19 | POLR2J | 16672 | 654 | 4.444 | 0.4105 | Yes | ||
| 20 | NUP107 | 8337 | 661 | 4.422 | 0.4267 | Yes | ||
| 21 | PSMD3 | 5803 | 692 | 4.290 | 0.4412 | Yes | ||
| 22 | PSMB5 | 9633 | 809 | 3.917 | 0.4496 | Yes | ||
| 23 | PSMD12 | 20621 | 821 | 3.878 | 0.4636 | Yes | ||
| 24 | NCBP2 | 12643 | 834 | 3.857 | 0.4774 | Yes | ||
| 25 | PSMA4 | 11179 | 882 | 3.710 | 0.4888 | Yes | ||
| 26 | PSMB6 | 9634 | 981 | 3.372 | 0.4961 | Yes | ||
| 27 | PSMD14 | 15005 | 1037 | 3.216 | 0.5052 | Yes | ||
| 28 | FEN1 | 8961 | 1068 | 3.138 | 0.5153 | Yes | ||
| 29 | PSMC2 | 16909 | 1164 | 2.940 | 0.5212 | Yes | ||
| 30 | PSME1 | 5300 9637 | 1201 | 2.861 | 0.5300 | Yes | ||
| 31 | PSMD9 | 3461 7393 | 1275 | 2.691 | 0.5362 | Yes | ||
| 32 | GTF2H2 | 6236 | 1365 | 2.487 | 0.5407 | Yes | ||
| 33 | TCEB1 | 13995 | 1381 | 2.456 | 0.5491 | Yes | ||
| 34 | TAF6 | 16322 891 | 1392 | 2.435 | 0.5577 | Yes | ||
| 35 | NUP62 | 9497 | 1445 | 2.341 | 0.5636 | Yes | ||
| 36 | AP2A2 | 18009 | 1448 | 2.337 | 0.5723 | Yes | ||
| 37 | GTF2F1 | 13599 | 1480 | 2.279 | 0.5792 | Yes | ||
| 38 | CCNH | 7322 | 1482 | 2.274 | 0.5877 | Yes | ||
| 39 | NUP37 | 3294 3326 19909 | 1523 | 2.230 | 0.5939 | Yes | ||
| 40 | POLR2I | 12839 | 1549 | 2.195 | 0.6007 | Yes | ||
| 41 | GTF2E2 | 18635 | 1569 | 2.165 | 0.6078 | Yes | ||
| 42 | RNMT | 7501 | 1694 | 1.984 | 0.6086 | Yes | ||
| 43 | NUP155 | 2298 5027 | 1713 | 1.943 | 0.6149 | Yes | ||
| 44 | GTF2E1 | 22597 | 1780 | 1.855 | 0.6183 | Yes | ||
| 45 | TH1L | 14715 | 1809 | 1.820 | 0.6236 | Yes | ||
| 46 | NUP160 | 14957 | 1842 | 1.755 | 0.6284 | Yes | ||
| 47 | PSMB3 | 11180 | 1849 | 1.748 | 0.6347 | Yes | ||
| 48 | PSMC1 | 9635 | 1857 | 1.738 | 0.6408 | Yes | ||
| 49 | RBX1 | 12130 7123 | 1911 | 1.661 | 0.6442 | Yes | ||
| 50 | TBP | 671 1554 | 1943 | 1.603 | 0.6485 | Yes | ||
| 51 | XRCC6 | 2286 8991 | 1950 | 1.597 | 0.6542 | Yes | ||
| 52 | MNAT1 | 9396 2161 | 2016 | 1.537 | 0.6564 | Yes | ||
| 53 | RANBP2 | 20019 | 2017 | 1.536 | 0.6622 | Yes | ||
| 54 | PSMA2 | 9631 | 2075 | 1.465 | 0.6646 | Yes | ||
| 55 | PSMC6 | 7379 | 2094 | 1.448 | 0.6691 | Yes | ||
| 56 | PSMA5 | 6464 | 2098 | 1.446 | 0.6743 | Yes | ||
| 57 | PACS1 | 23975 | 2128 | 1.400 | 0.6780 | Yes | ||
| 58 | POLR2E | 3325 19699 | 2210 | 1.298 | 0.6785 | Yes | ||
| 59 | ERCC3 | 23605 | 2238 | 1.255 | 0.6817 | Yes | ||
| 60 | PSMC3 | 9636 | 2250 | 1.240 | 0.6858 | Yes | ||
| 61 | PSMB4 | 15252 | 2297 | 1.181 | 0.6877 | Yes | ||
| 62 | TAF11 | 12733 | 2390 | 1.069 | 0.6868 | Yes | ||
| 63 | TAF5 | 23833 5934 3759 | 2443 | 1.030 | 0.6878 | Yes | ||
| 64 | PSMA3 | 9632 5298 | 2493 | 1.000 | 0.6889 | Yes | ||
| 65 | POLR2G | 23753 | 2511 | 0.997 | 0.6918 | Yes | ||
| 66 | POLR2A | 5394 | 2654 | 0.858 | 0.6873 | No | ||
| 67 | PSMD10 | 2578 2597 24046 | 2709 | 0.811 | 0.6874 | No | ||
| 68 | TCEB2 | 12563 23102 7476 | 2802 | 0.726 | 0.6852 | No | ||
| 69 | TAF12 | 2537 16058 2338 | 3042 | 0.532 | 0.6742 | No | ||
| 70 | KPNA1 | 22776 | 3097 | 0.498 | 0.6732 | No | ||
| 71 | RANGAP1 | 2180 22195 | 3162 | 0.457 | 0.6714 | No | ||
| 72 | SUPT4H1 | 9938 | 3948 | 0.222 | 0.6298 | No | ||
| 73 | GTF2H1 | 4069 18236 | 4080 | 0.203 | 0.6235 | No | ||
| 74 | TAF9 | 3213 8433 | 4594 | 0.141 | 0.5963 | No | ||
| 75 | PSMB1 | 23118 | 4750 | 0.128 | 0.5884 | No | ||
| 76 | PSMD4 | 15251 | 4763 | 0.127 | 0.5882 | No | ||
| 77 | FYN | 3375 3395 20052 | 5072 | 0.109 | 0.5720 | No | ||
| 78 | CD4 | 16999 | 5252 | 0.099 | 0.5627 | No | ||
| 79 | XPO1 | 4172 | 5363 | 0.093 | 0.5571 | No | ||
| 80 | XRCC5 | 14229 | 5838 | 0.072 | 0.5317 | No | ||
| 81 | RANBP1 | 9692 5357 | 5854 | 0.071 | 0.5312 | No | ||
| 82 | NUP35 | 12803 | 6597 | 0.051 | 0.4913 | No | ||
| 83 | GTF2A2 | 10654 | 6734 | 0.048 | 0.4841 | No | ||
| 84 | NUP188 | 15053 | 7801 | 0.028 | 0.4266 | No | ||
| 85 | PSMD5 | 2774 14612 | 7934 | 0.026 | 0.4195 | No | ||
| 86 | PSMA6 | 21270 | 8391 | 0.019 | 0.3949 | No | ||
| 87 | CD28 | 14239 4092 | 8632 | 0.016 | 0.3820 | No | ||
| 88 | TAF4 | 14319 | 8745 | 0.015 | 0.3760 | No | ||
| 89 | TPR | 927 4255 | 8987 | 0.012 | 0.3630 | No | ||
| 90 | B2M | 72 14882 | 9089 | 0.011 | 0.3576 | No | ||
| 91 | ERCC2 | 1549 18363 3812 | 9115 | 0.010 | 0.3563 | No | ||
| 92 | NUP153 | 21474 | 9407 | 0.006 | 0.3406 | No | ||
| 93 | CDK7 | 21365 | 9956 | -0.001 | 0.3110 | No | ||
| 94 | AP2B1 | 12958 7752 12957 1280 | 10881 | -0.012 | 0.2611 | No | ||
| 95 | GTF2A1 | 2064 8187 8188 13512 | 10949 | -0.013 | 0.2575 | No | ||
| 96 | XRCC4 | 4238 8432 | 10951 | -0.013 | 0.2575 | No | ||
| 97 | POLR2B | 16817 | 11867 | -0.028 | 0.2081 | No | ||
| 98 | PSMA1 | 1627 17669 | 12144 | -0.033 | 0.1933 | No | ||
| 99 | POLR2C | 9750 | 12998 | -0.054 | 0.1474 | No | ||
| 100 | PSMD11 | 12772 7600 | 14271 | -0.126 | 0.0791 | No | ||
| 101 | PPIA | 1284 11188 | 14937 | -0.225 | 0.0440 | No | ||
| 102 | LCK | 15746 | 15440 | -0.368 | 0.0182 | No | ||
| 103 | NUP54 | 11231 11232 6516 | 15488 | -0.387 | 0.0171 | No | ||
| 104 | RNGTT | 10769 2354 6272 | 15504 | -0.396 | 0.0178 | No | ||
| 105 | AP1G1 | 4392 | 15515 | -0.404 | 0.0188 | No | ||
| 106 | PAK2 | 10310 5875 | 15938 | -0.671 | -0.0015 | No | ||
| 107 | HCK | 14787 | 15979 | -0.695 | -0.0011 | No | ||
| 108 | LIG1 | 18388 1749 1493 | 16393 | -1.092 | -0.0193 | No | ||
| 109 | CCNT1 | 22140 11607 | 16429 | -1.133 | -0.0169 | No | ||
| 110 | GTF2B | 10489 | 16476 | -1.194 | -0.0149 | No | ||
| 111 | RAN | 5356 9691 | 16867 | -1.732 | -0.0295 | No | ||
| 112 | ELL | 18586 | 16902 | -1.772 | -0.0247 | No | ||
| 113 | AP1B1 | 8599 | 16957 | -1.842 | -0.0207 | No | ||
| 114 | CDK9 | 14619 | 16966 | -1.859 | -0.0142 | No | ||
| 115 | ARF1 | 64 | 17184 | -2.210 | -0.0176 | No | ||
| 116 | PSMB10 | 5299 18761 | 17214 | -2.264 | -0.0107 | No | ||
| 117 | TAF13 | 8288 | 17232 | -2.288 | -0.0030 | No | ||
| 118 | TAF10 | 10785 | 17301 | -2.423 | 0.0024 | No | ||
| 119 | TCEB3 | 15707 | 17985 | -4.177 | -0.0189 | No | ||
| 120 | LIG4 | 11454 | 18034 | -4.379 | -0.0051 | No | ||
| 121 | PSMB2 | 2324 16078 | 18611 | -9.723 | 0.0003 | No |