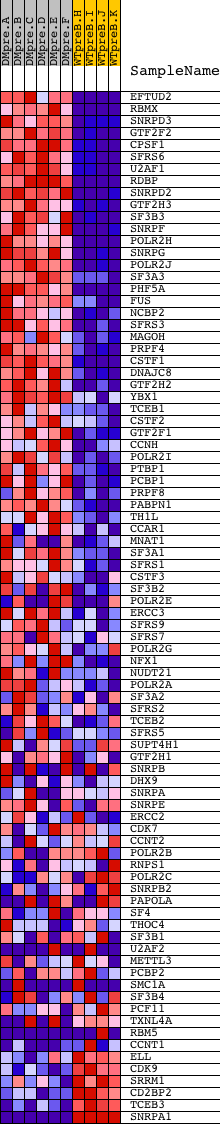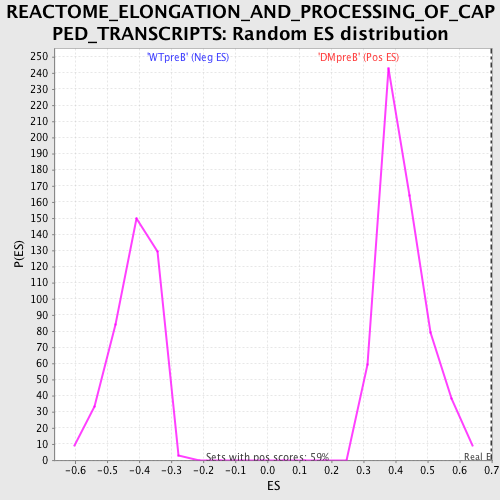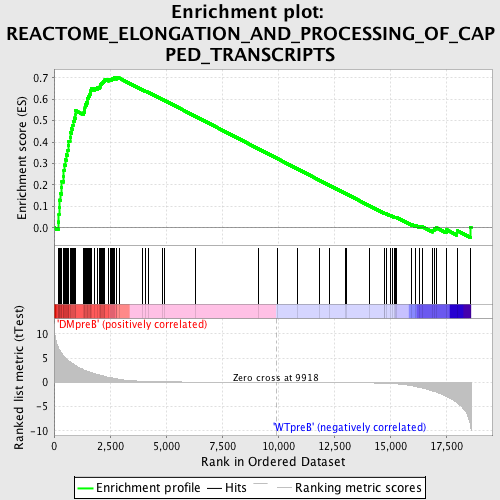
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | DMpreB |
| GeneSet | REACTOME_ELONGATION_AND_PROCESSING_OF_CAPPED_TRANSCRIPTS |
| Enrichment Score (ES) | 0.70104295 |
| Normalized Enrichment Score (NES) | 1.6570097 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.007850052 |
| FWER p-Value | 0.173 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | EFTUD2 | 1219 20203 | 183 | 7.206 | 0.0273 | Yes | ||
| 2 | RBMX | 5367 9708 | 214 | 6.907 | 0.0614 | Yes | ||
| 3 | SNRPD3 | 12514 | 225 | 6.797 | 0.0959 | Yes | ||
| 4 | GTF2F2 | 21750 | 236 | 6.685 | 0.1299 | Yes | ||
| 5 | CPSF1 | 22241 | 298 | 6.229 | 0.1587 | Yes | ||
| 6 | SFRS6 | 14751 | 326 | 6.012 | 0.1883 | Yes | ||
| 7 | U2AF1 | 8431 | 346 | 5.954 | 0.2180 | Yes | ||
| 8 | RDBP | 6589 | 428 | 5.370 | 0.2414 | Yes | ||
| 9 | SNRPD2 | 8412 | 436 | 5.355 | 0.2686 | Yes | ||
| 10 | GTF2H3 | 5551 | 477 | 5.199 | 0.2933 | Yes | ||
| 11 | SF3B3 | 18746 | 507 | 5.015 | 0.3176 | Yes | ||
| 12 | SNRPF | 7645 | 541 | 4.871 | 0.3410 | Yes | ||
| 13 | POLR2H | 10888 | 611 | 4.591 | 0.3610 | Yes | ||
| 14 | SNRPG | 12622 | 632 | 4.487 | 0.3830 | Yes | ||
| 15 | POLR2J | 16672 | 654 | 4.444 | 0.4049 | Yes | ||
| 16 | SF3A3 | 16091 | 711 | 4.229 | 0.4237 | Yes | ||
| 17 | PHF5A | 22194 | 738 | 4.141 | 0.4436 | Yes | ||
| 18 | FUS | 6135 | 784 | 3.989 | 0.4618 | Yes | ||
| 19 | NCBP2 | 12643 | 834 | 3.857 | 0.4791 | Yes | ||
| 20 | SFRS3 | 5428 23312 | 850 | 3.817 | 0.4980 | Yes | ||
| 21 | MAGOH | 2386 16148 | 921 | 3.574 | 0.5126 | Yes | ||
| 22 | PRPF4 | 7655 12845 | 949 | 3.476 | 0.5291 | Yes | ||
| 23 | CSTF1 | 12515 | 954 | 3.455 | 0.5467 | Yes | ||
| 24 | DNAJC8 | 12712 | 1329 | 2.559 | 0.5398 | Yes | ||
| 25 | GTF2H2 | 6236 | 1365 | 2.487 | 0.5507 | Yes | ||
| 26 | YBX1 | 5929 2389 | 1376 | 2.461 | 0.5629 | Yes | ||
| 27 | TCEB1 | 13995 | 1381 | 2.456 | 0.5753 | Yes | ||
| 28 | CSTF2 | 24257 | 1440 | 2.349 | 0.5843 | Yes | ||
| 29 | GTF2F1 | 13599 | 1480 | 2.279 | 0.5940 | Yes | ||
| 30 | CCNH | 7322 | 1482 | 2.274 | 0.6057 | Yes | ||
| 31 | POLR2I | 12839 | 1549 | 2.195 | 0.6134 | Yes | ||
| 32 | PTBP1 | 5303 | 1596 | 2.132 | 0.6220 | Yes | ||
| 33 | PCBP1 | 17085 | 1616 | 2.102 | 0.6318 | Yes | ||
| 34 | PRPF8 | 20780 1371 | 1640 | 2.062 | 0.6412 | Yes | ||
| 35 | PABPN1 | 12018 | 1676 | 2.019 | 0.6497 | Yes | ||
| 36 | TH1L | 14715 | 1809 | 1.820 | 0.6520 | Yes | ||
| 37 | CCAR1 | 7454 | 1939 | 1.606 | 0.6533 | Yes | ||
| 38 | MNAT1 | 9396 2161 | 2016 | 1.537 | 0.6572 | Yes | ||
| 39 | SF3A1 | 7450 | 2053 | 1.492 | 0.6629 | Yes | ||
| 40 | SFRS1 | 8492 | 2072 | 1.466 | 0.6695 | Yes | ||
| 41 | CSTF3 | 10449 6002 | 2125 | 1.404 | 0.6740 | Yes | ||
| 42 | SF3B2 | 23974 | 2176 | 1.345 | 0.6782 | Yes | ||
| 43 | POLR2E | 3325 19699 | 2210 | 1.298 | 0.6831 | Yes | ||
| 44 | ERCC3 | 23605 | 2238 | 1.255 | 0.6881 | Yes | ||
| 45 | SFRS9 | 16731 | 2268 | 1.221 | 0.6929 | Yes | ||
| 46 | SFRS7 | 22889 | 2416 | 1.053 | 0.6904 | Yes | ||
| 47 | POLR2G | 23753 | 2511 | 0.997 | 0.6904 | Yes | ||
| 48 | NFX1 | 2475 | 2552 | 0.972 | 0.6933 | Yes | ||
| 49 | NUDT21 | 12665 | 2612 | 0.905 | 0.6948 | Yes | ||
| 50 | POLR2A | 5394 | 2654 | 0.858 | 0.6970 | Yes | ||
| 51 | SF3A2 | 19938 | 2677 | 0.837 | 0.7001 | Yes | ||
| 52 | SFRS2 | 9807 20136 | 2789 | 0.733 | 0.6979 | Yes | ||
| 53 | TCEB2 | 12563 23102 7476 | 2802 | 0.726 | 0.7010 | Yes | ||
| 54 | SFRS5 | 9808 2062 | 2896 | 0.639 | 0.6993 | No | ||
| 55 | SUPT4H1 | 9938 | 3948 | 0.222 | 0.6438 | No | ||
| 56 | GTF2H1 | 4069 18236 | 4080 | 0.203 | 0.6377 | No | ||
| 57 | SNRPB | 9842 5469 2736 | 4208 | 0.184 | 0.6318 | No | ||
| 58 | DHX9 | 8845 4608 3948 | 4843 | 0.122 | 0.5982 | No | ||
| 59 | SNRPA | 7007 11992 7008 | 4916 | 0.117 | 0.5950 | No | ||
| 60 | SNRPE | 9843 | 6295 | 0.058 | 0.5209 | No | ||
| 61 | ERCC2 | 1549 18363 3812 | 9115 | 0.010 | 0.3688 | No | ||
| 62 | CDK7 | 21365 | 9956 | -0.001 | 0.3235 | No | ||
| 63 | CCNT2 | 3993 13079 | 10873 | -0.012 | 0.2741 | No | ||
| 64 | POLR2B | 16817 | 11867 | -0.028 | 0.2207 | No | ||
| 65 | RNPS1 | 9730 23361 | 12295 | -0.036 | 0.1978 | No | ||
| 66 | POLR2C | 9750 | 12998 | -0.054 | 0.1602 | No | ||
| 67 | SNRPB2 | 5470 | 13068 | -0.056 | 0.1568 | No | ||
| 68 | PAPOLA | 5261 9585 | 14063 | -0.107 | 0.1037 | No | ||
| 69 | SF4 | 18593 | 14746 | -0.191 | 0.0679 | No | ||
| 70 | THOC4 | 5710 10114 | 14757 | -0.194 | 0.0684 | No | ||
| 71 | SF3B1 | 13954 | 14840 | -0.208 | 0.0650 | No | ||
| 72 | U2AF2 | 5813 5812 5814 | 15014 | -0.242 | 0.0569 | No | ||
| 73 | METTL3 | 21841 | 15104 | -0.265 | 0.0535 | No | ||
| 74 | PCBP2 | 9533 | 15213 | -0.292 | 0.0492 | No | ||
| 75 | SMC1A | 2572 24225 6279 10780 | 15233 | -0.298 | 0.0497 | No | ||
| 76 | SF3B4 | 22269 | 15268 | -0.309 | 0.0494 | No | ||
| 77 | PCF11 | 121 | 15957 | -0.683 | 0.0158 | No | ||
| 78 | TXNL4A | 6567 11329 | 16111 | -0.840 | 0.0119 | No | ||
| 79 | RBM5 | 13507 3016 | 16304 | -1.004 | 0.0067 | No | ||
| 80 | CCNT1 | 22140 11607 | 16429 | -1.133 | 0.0059 | No | ||
| 81 | ELL | 18586 | 16902 | -1.772 | -0.0104 | No | ||
| 82 | CDK9 | 14619 | 16966 | -1.859 | -0.0042 | No | ||
| 83 | SRRM1 | 6958 | 17048 | -1.978 | 0.0016 | No | ||
| 84 | CD2BP2 | 17621 | 17506 | -2.877 | -0.0082 | No | ||
| 85 | TCEB3 | 15707 | 17985 | -4.177 | -0.0124 | No | ||
| 86 | SNRPA1 | 1373 18216 | 18588 | -8.994 | 0.0015 | No |