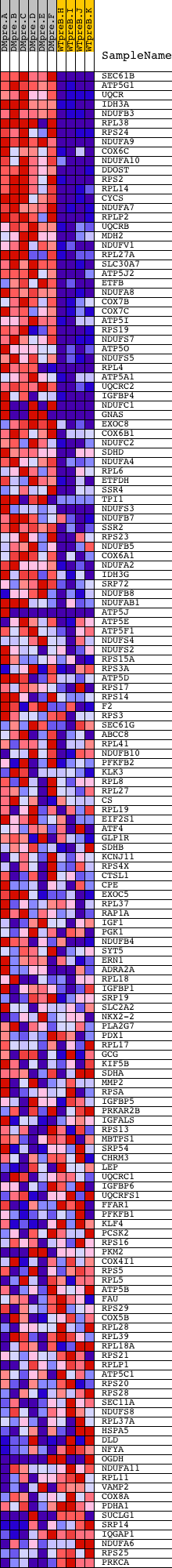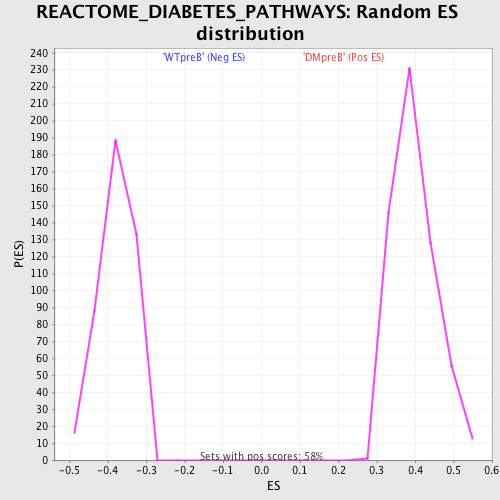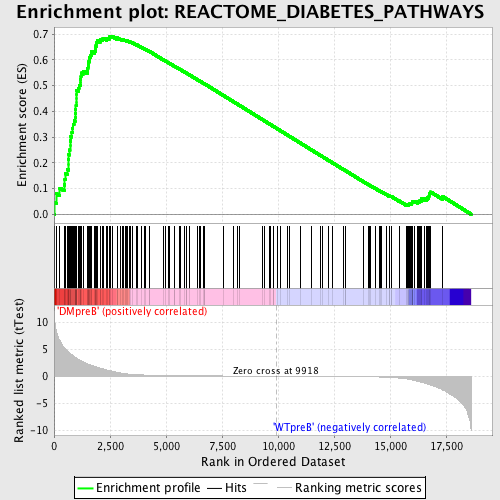
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | DMpreB |
| GeneSet | REACTOME_DIABETES_PATHWAYS |
| Enrichment Score (ES) | 0.69221216 |
| Normalized Enrichment Score (NES) | 1.7433953 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0013953076 |
| FWER p-Value | 0.01 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SEC61B | 12305 7266 | 18 | 10.626 | 0.0468 | Yes | ||
| 2 | ATP5G1 | 8636 | 95 | 8.444 | 0.0806 | Yes | ||
| 3 | UQCR | 12382 | 255 | 6.585 | 0.1015 | Yes | ||
| 4 | IDH3A | 19447 | 451 | 5.296 | 0.1148 | Yes | ||
| 5 | NDUFB3 | 12362 | 480 | 5.178 | 0.1365 | Yes | ||
| 6 | RPL38 | 12562 20606 7475 | 515 | 4.983 | 0.1571 | Yes | ||
| 7 | RPS24 | 5399 | 584 | 4.691 | 0.1744 | Yes | ||
| 8 | NDUFA9 | 12288 | 639 | 4.471 | 0.1916 | Yes | ||
| 9 | COX6C | 8775 | 641 | 4.467 | 0.2116 | Yes | ||
| 10 | NDUFA10 | 7420 | 655 | 4.441 | 0.2309 | Yes | ||
| 11 | DDOST | 16021 | 671 | 4.396 | 0.2498 | Yes | ||
| 12 | RPS2 | 9279 | 710 | 4.232 | 0.2668 | Yes | ||
| 13 | RPL14 | 19267 | 713 | 4.224 | 0.2856 | Yes | ||
| 14 | CYCS | 8821 | 744 | 4.120 | 0.3025 | Yes | ||
| 15 | NDUFA7 | 12347 | 793 | 3.955 | 0.3177 | Yes | ||
| 16 | RPLP2 | 7401 | 825 | 3.870 | 0.3334 | Yes | ||
| 17 | UQCRB | 12545 | 843 | 3.837 | 0.3497 | Yes | ||
| 18 | MDH2 | 9399 | 890 | 3.693 | 0.3638 | Yes | ||
| 19 | NDUFV1 | 23953 | 938 | 3.511 | 0.3770 | Yes | ||
| 20 | RPL27A | 11181 6467 18130 | 948 | 3.476 | 0.3922 | Yes | ||
| 21 | SLC30A7 | 15186 | 965 | 3.430 | 0.4067 | Yes | ||
| 22 | ATP5J2 | 12186 | 976 | 3.393 | 0.4214 | Yes | ||
| 23 | ETFB | 18279 | 988 | 3.350 | 0.4359 | Yes | ||
| 24 | NDUFA8 | 14605 | 990 | 3.349 | 0.4508 | Yes | ||
| 25 | COX7B | 7260 | 992 | 3.337 | 0.4658 | Yes | ||
| 26 | COX7C | 8776 | 997 | 3.325 | 0.4805 | Yes | ||
| 27 | ATP5I | 8637 | 1084 | 3.109 | 0.4898 | Yes | ||
| 28 | RPS19 | 5398 | 1114 | 3.041 | 0.5019 | Yes | ||
| 29 | NDUFS7 | 19946 | 1179 | 2.911 | 0.5115 | Yes | ||
| 30 | ATP5O | 22539 | 1183 | 2.898 | 0.5244 | Yes | ||
| 31 | NDUFS5 | 9296 | 1198 | 2.871 | 0.5365 | Yes | ||
| 32 | RPL4 | 7499 19411 | 1205 | 2.853 | 0.5490 | Yes | ||
| 33 | ATP5A1 | 23505 | 1292 | 2.659 | 0.5563 | Yes | ||
| 34 | UQCRC2 | 18102 | 1481 | 2.276 | 0.5563 | Yes | ||
| 35 | IGFBP4 | 20670 9160 | 1497 | 2.250 | 0.5656 | Yes | ||
| 36 | NDUFC1 | 7287 12339 | 1514 | 2.233 | 0.5748 | Yes | ||
| 37 | GNAS | 9025 2963 2752 | 1524 | 2.229 | 0.5843 | Yes | ||
| 38 | EXOC8 | 4161 | 1550 | 2.193 | 0.5928 | Yes | ||
| 39 | COX6B1 | 4283 8479 | 1574 | 2.159 | 0.6013 | Yes | ||
| 40 | NDUFC2 | 18183 | 1590 | 2.139 | 0.6100 | Yes | ||
| 41 | SDHD | 19125 | 1637 | 2.073 | 0.6169 | Yes | ||
| 42 | NDUFA4 | 9451 | 1660 | 2.041 | 0.6248 | Yes | ||
| 43 | RPL6 | 9747 | 1668 | 2.029 | 0.6336 | Yes | ||
| 44 | ETFDH | 15313 | 1808 | 1.822 | 0.6342 | Yes | ||
| 45 | SSR4 | 24303 5519 | 1852 | 1.745 | 0.6397 | Yes | ||
| 46 | TPI1 | 5795 10212 | 1858 | 1.737 | 0.6473 | Yes | ||
| 47 | NDUFS3 | 7553 12683 | 1866 | 1.729 | 0.6547 | Yes | ||
| 48 | NDUFB7 | 12429 | 1892 | 1.682 | 0.6609 | Yes | ||
| 49 | SSR2 | 15549 | 1905 | 1.667 | 0.6677 | Yes | ||
| 50 | RPS23 | 12352 | 1923 | 1.642 | 0.6742 | Yes | ||
| 51 | NDUFB5 | 12277 | 2052 | 1.493 | 0.6739 | Yes | ||
| 52 | COX6A1 | 8774 4553 | 2066 | 1.470 | 0.6798 | Yes | ||
| 53 | NDUFA2 | 9450 | 2156 | 1.374 | 0.6812 | Yes | ||
| 54 | IDH3G | 24132 | 2215 | 1.290 | 0.6838 | Yes | ||
| 55 | SRP72 | 16819 | 2354 | 1.113 | 0.6814 | Yes | ||
| 56 | NDUFB8 | 7418 | 2404 | 1.062 | 0.6835 | Yes | ||
| 57 | NDUFAB1 | 7667 | 2456 | 1.022 | 0.6853 | Yes | ||
| 58 | ATP5J | 880 22555 | 2469 | 1.011 | 0.6892 | Yes | ||
| 59 | ATP5E | 14321 | 2497 | 0.999 | 0.6922 | Yes | ||
| 60 | ATP5F1 | 15212 | 2606 | 0.908 | 0.6904 | No | ||
| 61 | NDUFS4 | 9452 5157 3173 | 2810 | 0.721 | 0.6827 | No | ||
| 62 | NDUFS2 | 4089 13760 | 2812 | 0.719 | 0.6859 | No | ||
| 63 | RPS15A | 6476 | 2968 | 0.585 | 0.6801 | No | ||
| 64 | RPS3A | 9755 | 3073 | 0.515 | 0.6768 | No | ||
| 65 | ATP5D | 19949 | 3095 | 0.499 | 0.6779 | No | ||
| 66 | RPS17 | 9753 | 3181 | 0.449 | 0.6753 | No | ||
| 67 | RPS14 | 9751 | 3235 | 0.427 | 0.6743 | No | ||
| 68 | F2 | 14524 | 3282 | 0.398 | 0.6736 | No | ||
| 69 | RPS3 | 6549 11295 | 3374 | 0.362 | 0.6703 | No | ||
| 70 | SEC61G | 9795 | 3425 | 0.347 | 0.6692 | No | ||
| 71 | ABCC8 | 9941 11649 | 3509 | 0.324 | 0.6661 | No | ||
| 72 | RPL41 | 12611 | 3691 | 0.275 | 0.6576 | No | ||
| 73 | NDUFB10 | 23084 | 3707 | 0.271 | 0.6580 | No | ||
| 74 | PFKFB2 | 4116 5242 | 3903 | 0.230 | 0.6484 | No | ||
| 75 | KLK3 | 16337 | 4041 | 0.208 | 0.6419 | No | ||
| 76 | RPL8 | 22437 | 4060 | 0.205 | 0.6419 | No | ||
| 77 | RPL27 | 9740 | 4251 | 0.178 | 0.6324 | No | ||
| 78 | CS | 19839 | 4271 | 0.177 | 0.6322 | No | ||
| 79 | RPL19 | 9736 | 4895 | 0.119 | 0.5989 | No | ||
| 80 | EIF2S1 | 4658 | 4969 | 0.114 | 0.5955 | No | ||
| 81 | ATF4 | 22417 2239 | 5109 | 0.106 | 0.5884 | No | ||
| 82 | GLP1R | 23304 | 5165 | 0.103 | 0.5859 | No | ||
| 83 | SDHB | 2348 12566 14880 | 5368 | 0.092 | 0.5754 | No | ||
| 84 | KCNJ11 | 17822 | 5383 | 0.091 | 0.5751 | No | ||
| 85 | RPS4X | 9756 | 5613 | 0.081 | 0.5630 | No | ||
| 86 | CTSL1 | 4574 | 5643 | 0.079 | 0.5618 | No | ||
| 87 | CPE | 18868 | 5831 | 0.072 | 0.5520 | No | ||
| 88 | EXOC5 | 8370 | 5898 | 0.070 | 0.5487 | No | ||
| 89 | RPL37 | 12502 7421 22521 | 5901 | 0.070 | 0.5489 | No | ||
| 90 | RAP1A | 8467 | 6027 | 0.066 | 0.5425 | No | ||
| 91 | IGF1 | 3352 9156 3409 | 6411 | 0.055 | 0.5220 | No | ||
| 92 | PGK1 | 9557 5244 | 6483 | 0.054 | 0.5183 | No | ||
| 93 | NDUFB4 | 22596 7541 | 6523 | 0.053 | 0.5165 | No | ||
| 94 | SYT5 | 17986 | 6685 | 0.049 | 0.5080 | No | ||
| 95 | ERN1 | 8137 | 6692 | 0.049 | 0.5079 | No | ||
| 96 | ADRA2A | 4355 | 6721 | 0.048 | 0.5066 | No | ||
| 97 | RPL18 | 450 5390 | 7540 | 0.032 | 0.4624 | No | ||
| 98 | IGFBP1 | 20948 | 7987 | 0.025 | 0.4383 | No | ||
| 99 | SRP19 | 2004 12341 | 8165 | 0.022 | 0.4289 | No | ||
| 100 | SLC2A2 | 15623 1880 1855 | 8275 | 0.021 | 0.4230 | No | ||
| 101 | NKX2-2 | 5176 | 9308 | 0.008 | 0.3672 | No | ||
| 102 | PLA2G7 | 23233 | 9373 | 0.007 | 0.3637 | No | ||
| 103 | PDX1 | 16621 | 9626 | 0.004 | 0.3501 | No | ||
| 104 | RPL17 | 11429 6653 | 9669 | 0.003 | 0.3478 | No | ||
| 105 | GCG | 14578 | 9789 | 0.002 | 0.3414 | No | ||
| 106 | KIF5B | 4955 2031 | 9793 | 0.002 | 0.3412 | No | ||
| 107 | SDHA | 12436 | 9969 | -0.001 | 0.3318 | No | ||
| 108 | MMP2 | 18525 | 10110 | -0.002 | 0.3242 | No | ||
| 109 | RPSA | 19270 4984 | 10413 | -0.006 | 0.3078 | No | ||
| 110 | IGFBP5 | 4899 | 10506 | -0.007 | 0.3029 | No | ||
| 111 | PRKAR2B | 5288 2107 | 11006 | -0.014 | 0.2759 | No | ||
| 112 | IGFALS | 9158 | 11488 | -0.021 | 0.2500 | No | ||
| 113 | RPS13 | 12633 | 11876 | -0.028 | 0.2291 | No | ||
| 114 | MBTPS1 | 18730 3809 3770 | 11960 | -0.029 | 0.2248 | No | ||
| 115 | SRP54 | 6282 | 12262 | -0.036 | 0.2086 | No | ||
| 116 | CHRM3 | 21547 | 12439 | -0.039 | 0.1993 | No | ||
| 117 | LEP | 17505 | 12936 | -0.052 | 0.1726 | No | ||
| 118 | UQCRC1 | 10259 | 12988 | -0.054 | 0.1701 | No | ||
| 119 | IGFBP6 | 9161 | 13028 | -0.055 | 0.1682 | No | ||
| 120 | UQCRFS1 | 21499 | 13797 | -0.089 | 0.1270 | No | ||
| 121 | FFAR1 | 10583 | 14031 | -0.104 | 0.1149 | No | ||
| 122 | PFKFB1 | 9552 | 14062 | -0.107 | 0.1137 | No | ||
| 123 | KLF4 | 9230 4962 4963 | 14142 | -0.113 | 0.1099 | No | ||
| 124 | PCSK2 | 9537 5231 14824 | 14347 | -0.135 | 0.0995 | No | ||
| 125 | RPS16 | 9752 | 14531 | -0.155 | 0.0903 | No | ||
| 126 | PKM2 | 3642 9573 | 14550 | -0.157 | 0.0900 | No | ||
| 127 | COX4I1 | 18444 | 14624 | -0.170 | 0.0868 | No | ||
| 128 | RPS5 | 18391 | 14818 | -0.203 | 0.0773 | No | ||
| 129 | RPL5 | 9746 | 14950 | -0.227 | 0.0712 | No | ||
| 130 | ATP5B | 19846 | 14988 | -0.238 | 0.0703 | No | ||
| 131 | FAU | 8954 | 15042 | -0.247 | 0.0685 | No | ||
| 132 | RPS29 | 9754 | 15047 | -0.248 | 0.0694 | No | ||
| 133 | COX5B | 4552 | 15403 | -0.353 | 0.0518 | No | ||
| 134 | RPL28 | 5392 | 15723 | -0.504 | 0.0367 | No | ||
| 135 | RPL39 | 12496 | 15794 | -0.545 | 0.0354 | No | ||
| 136 | RPL18A | 13358 | 15815 | -0.562 | 0.0368 | No | ||
| 137 | RPS21 | 12356 | 15832 | -0.577 | 0.0386 | No | ||
| 138 | RPLP1 | 3010 | 15852 | -0.591 | 0.0402 | No | ||
| 139 | ATP5C1 | 8635 | 15887 | -0.627 | 0.0412 | No | ||
| 140 | RPS20 | 7438 | 15960 | -0.687 | 0.0403 | No | ||
| 141 | RPS28 | 12009 | 15976 | -0.693 | 0.0426 | No | ||
| 142 | SEC11A | 12144 | 15977 | -0.693 | 0.0458 | No | ||
| 143 | NDUFS8 | 23950 | 15985 | -0.705 | 0.0485 | No | ||
| 144 | RPL37A | 9744 | 15994 | -0.712 | 0.0513 | No | ||
| 145 | HSPA5 | 9045 | 16089 | -0.811 | 0.0499 | No | ||
| 146 | DLD | 2097 21090 | 16216 | -0.942 | 0.0473 | No | ||
| 147 | NFYA | 5172 | 16265 | -0.988 | 0.0491 | No | ||
| 148 | OGDH | 9502 1412 | 16302 | -1.003 | 0.0517 | No | ||
| 149 | NDUFA11 | 12832 | 16348 | -1.042 | 0.0539 | No | ||
| 150 | RPL11 | 12450 | 16403 | -1.103 | 0.0559 | No | ||
| 151 | VAMP2 | 20826 | 16407 | -1.104 | 0.0607 | No | ||
| 152 | COX8A | 8777 | 16528 | -1.248 | 0.0598 | No | ||
| 153 | PDHA1 | 24020 | 16629 | -1.404 | 0.0607 | No | ||
| 154 | SUCLG1 | 17409 1059 | 16678 | -1.463 | 0.0647 | No | ||
| 155 | SRP14 | 9894 | 16731 | -1.553 | 0.0689 | No | ||
| 156 | IQGAP1 | 6619 | 16738 | -1.569 | 0.0756 | No | ||
| 157 | NDUFA6 | 12473 | 16746 | -1.581 | 0.0823 | No | ||
| 158 | RPS25 | 13270 | 16809 | -1.680 | 0.0865 | No | ||
| 159 | PRKCA | 20174 | 17337 | -2.525 | 0.0693 | No |