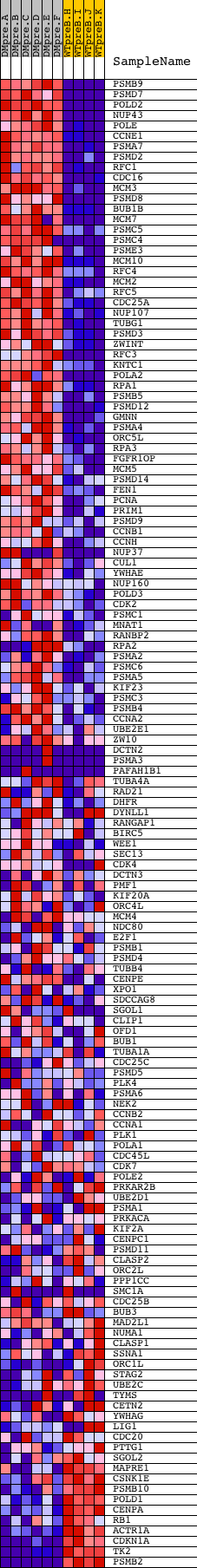
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | DMpreB |
| GeneSet | REACTOME_CELL_CYCLE__MITOTIC |
| Enrichment Score (ES) | 0.6672519 |
| Normalized Enrichment Score (NES) | 1.6579976 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.00799658 |
| FWER p-Value | 0.168 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSMB9 | 23021 | 15 | 10.811 | 0.0338 | Yes | ||
| 2 | PSMD7 | 18752 3825 | 28 | 10.182 | 0.0657 | Yes | ||
| 3 | POLD2 | 20537 | 74 | 8.897 | 0.0917 | Yes | ||
| 4 | NUP43 | 20094 | 90 | 8.530 | 0.1182 | Yes | ||
| 5 | POLE | 16755 | 244 | 6.625 | 0.1311 | Yes | ||
| 6 | CCNE1 | 17857 | 261 | 6.545 | 0.1511 | Yes | ||
| 7 | PSMA7 | 14318 | 278 | 6.350 | 0.1706 | Yes | ||
| 8 | PSMD2 | 10137 5724 | 288 | 6.286 | 0.1902 | Yes | ||
| 9 | RFC1 | 16527 | 335 | 5.989 | 0.2068 | Yes | ||
| 10 | CDC16 | 18676 | 340 | 5.977 | 0.2257 | Yes | ||
| 11 | MCM3 | 13991 | 390 | 5.583 | 0.2409 | Yes | ||
| 12 | PSMD8 | 7166 | 418 | 5.413 | 0.2568 | Yes | ||
| 13 | BUB1B | 14908 | 474 | 5.208 | 0.2705 | Yes | ||
| 14 | MCM7 | 9372 3568 | 497 | 5.089 | 0.2855 | Yes | ||
| 15 | PSMC5 | 1348 20624 | 522 | 4.954 | 0.3001 | Yes | ||
| 16 | PSMC4 | 2141 17915 | 531 | 4.907 | 0.3154 | Yes | ||
| 17 | PSME3 | 20657 | 553 | 4.832 | 0.3297 | Yes | ||
| 18 | MCM10 | 14694 | 573 | 4.728 | 0.3438 | Yes | ||
| 19 | RFC4 | 1735 22627 | 589 | 4.674 | 0.3579 | Yes | ||
| 20 | MCM2 | 17074 | 598 | 4.627 | 0.3723 | Yes | ||
| 21 | RFC5 | 13005 7791 | 625 | 4.526 | 0.3853 | Yes | ||
| 22 | CDC25A | 8721 | 631 | 4.494 | 0.3994 | Yes | ||
| 23 | NUP107 | 8337 | 661 | 4.422 | 0.4120 | Yes | ||
| 24 | TUBG1 | 20662 | 674 | 4.381 | 0.4254 | Yes | ||
| 25 | PSMD3 | 5803 | 692 | 4.290 | 0.4382 | Yes | ||
| 26 | ZWINT | 19992 | 701 | 4.269 | 0.4514 | Yes | ||
| 27 | RFC3 | 12786 | 722 | 4.193 | 0.4637 | Yes | ||
| 28 | KNTC1 | 16705 | 725 | 4.189 | 0.4770 | Yes | ||
| 29 | POLA2 | 23988 | 733 | 4.174 | 0.4900 | Yes | ||
| 30 | RPA1 | 20349 | 739 | 4.140 | 0.5029 | Yes | ||
| 31 | PSMB5 | 9633 | 809 | 3.917 | 0.5117 | Yes | ||
| 32 | PSMD12 | 20621 | 821 | 3.878 | 0.5235 | Yes | ||
| 33 | GMNN | 21513 | 848 | 3.827 | 0.5344 | Yes | ||
| 34 | PSMA4 | 11179 | 882 | 3.710 | 0.5444 | Yes | ||
| 35 | ORC5L | 11173 3595 | 924 | 3.572 | 0.5536 | Yes | ||
| 36 | RPA3 | 12667 | 963 | 3.441 | 0.5626 | Yes | ||
| 37 | FGFR1OP | 23391 | 996 | 3.325 | 0.5715 | Yes | ||
| 38 | MCM5 | 18564 | 1005 | 3.303 | 0.5816 | Yes | ||
| 39 | PSMD14 | 15005 | 1037 | 3.216 | 0.5902 | Yes | ||
| 40 | FEN1 | 8961 | 1068 | 3.138 | 0.5986 | Yes | ||
| 41 | PCNA | 9535 | 1100 | 3.074 | 0.6068 | Yes | ||
| 42 | PRIM1 | 19847 | 1169 | 2.930 | 0.6125 | Yes | ||
| 43 | PSMD9 | 3461 7393 | 1275 | 2.691 | 0.6154 | Yes | ||
| 44 | CCNB1 | 11201 21362 | 1458 | 2.324 | 0.6130 | Yes | ||
| 45 | CCNH | 7322 | 1482 | 2.274 | 0.6190 | Yes | ||
| 46 | NUP37 | 3294 3326 19909 | 1523 | 2.230 | 0.6240 | Yes | ||
| 47 | CUL1 | 17462 | 1686 | 2.000 | 0.6216 | Yes | ||
| 48 | YWHAE | 20776 | 1727 | 1.923 | 0.6256 | Yes | ||
| 49 | NUP160 | 14957 | 1842 | 1.755 | 0.6250 | Yes | ||
| 50 | POLD3 | 17742 | 1845 | 1.754 | 0.6305 | Yes | ||
| 51 | CDK2 | 3438 3373 19592 3322 | 1846 | 1.753 | 0.6361 | Yes | ||
| 52 | PSMC1 | 9635 | 1857 | 1.738 | 0.6412 | Yes | ||
| 53 | MNAT1 | 9396 2161 | 2016 | 1.537 | 0.6375 | Yes | ||
| 54 | RANBP2 | 20019 | 2017 | 1.536 | 0.6424 | Yes | ||
| 55 | RPA2 | 2330 16057 | 2021 | 1.531 | 0.6472 | Yes | ||
| 56 | PSMA2 | 9631 | 2075 | 1.465 | 0.6490 | Yes | ||
| 57 | PSMC6 | 7379 | 2094 | 1.448 | 0.6526 | Yes | ||
| 58 | PSMA5 | 6464 | 2098 | 1.446 | 0.6571 | Yes | ||
| 59 | KIF23 | 19091 | 2101 | 1.442 | 0.6616 | Yes | ||
| 60 | PSMC3 | 9636 | 2250 | 1.240 | 0.6575 | Yes | ||
| 61 | PSMB4 | 15252 | 2297 | 1.181 | 0.6588 | Yes | ||
| 62 | CCNA2 | 15357 | 2301 | 1.176 | 0.6624 | Yes | ||
| 63 | UBE2E1 | 10243 5817 | 2423 | 1.047 | 0.6592 | Yes | ||
| 64 | ZW10 | 3114 19464 | 2458 | 1.020 | 0.6607 | Yes | ||
| 65 | DCTN2 | 7635 12812 | 2462 | 1.017 | 0.6637 | Yes | ||
| 66 | PSMA3 | 9632 5298 | 2493 | 1.000 | 0.6653 | Yes | ||
| 67 | PAFAH1B1 | 1340 5220 9524 | 2517 | 0.994 | 0.6673 | Yes | ||
| 68 | TUBA4A | 10232 | 2723 | 0.792 | 0.6587 | No | ||
| 69 | RAD21 | 22298 | 2816 | 0.716 | 0.6560 | No | ||
| 70 | DHFR | 21590 | 2941 | 0.609 | 0.6512 | No | ||
| 71 | DYNLL1 | 12135 | 3037 | 0.535 | 0.6478 | No | ||
| 72 | RANGAP1 | 2180 22195 | 3162 | 0.457 | 0.6426 | No | ||
| 73 | BIRC5 | 8611 8610 4399 | 3195 | 0.443 | 0.6422 | No | ||
| 74 | WEE1 | 18127 | 3229 | 0.429 | 0.6418 | No | ||
| 75 | SEC13 | 17039 | 3251 | 0.416 | 0.6420 | No | ||
| 76 | CDK4 | 3424 19859 | 3277 | 0.401 | 0.6419 | No | ||
| 77 | DCTN3 | 15908 | 3733 | 0.266 | 0.6182 | No | ||
| 78 | PMF1 | 12452 | 3800 | 0.251 | 0.6154 | No | ||
| 79 | KIF20A | 23602 | 4017 | 0.212 | 0.6044 | No | ||
| 80 | ORC4L | 11172 6460 | 4035 | 0.208 | 0.6041 | No | ||
| 81 | MCM4 | 22655 1708 | 4196 | 0.186 | 0.5961 | No | ||
| 82 | NDC80 | 22900 | 4402 | 0.160 | 0.5855 | No | ||
| 83 | E2F1 | 14384 | 4565 | 0.144 | 0.5772 | No | ||
| 84 | PSMB1 | 23118 | 4750 | 0.128 | 0.5676 | No | ||
| 85 | PSMD4 | 15251 | 4763 | 0.127 | 0.5674 | No | ||
| 86 | TUBB4 | 22917 | 4849 | 0.121 | 0.5632 | No | ||
| 87 | CENPE | 15414 | 5351 | 0.093 | 0.5363 | No | ||
| 88 | XPO1 | 4172 | 5363 | 0.093 | 0.5360 | No | ||
| 89 | SDCCAG8 | 14034 | 5385 | 0.091 | 0.5352 | No | ||
| 90 | SGOL1 | 7810 7809 13030 | 5954 | 0.068 | 0.5047 | No | ||
| 91 | CLIP1 | 3603 7121 | 6041 | 0.065 | 0.5002 | No | ||
| 92 | OFD1 | 24007 | 6434 | 0.055 | 0.4792 | No | ||
| 93 | BUB1 | 8665 | 7337 | 0.036 | 0.4305 | No | ||
| 94 | TUBA1A | 1130 10233 5807 5806 | 7444 | 0.034 | 0.4248 | No | ||
| 95 | CDC25C | 23468 1954 1977 | 7624 | 0.031 | 0.4152 | No | ||
| 96 | PSMD5 | 2774 14612 | 7934 | 0.026 | 0.3986 | No | ||
| 97 | PLK4 | 1870 1874 | 8253 | 0.021 | 0.3815 | No | ||
| 98 | PSMA6 | 21270 | 8391 | 0.019 | 0.3741 | No | ||
| 99 | NEK2 | 5160 | 9198 | 0.009 | 0.3305 | No | ||
| 100 | CCNB2 | 19067 | 9366 | 0.007 | 0.3215 | No | ||
| 101 | CCNA1 | 15340 | 9443 | 0.006 | 0.3174 | No | ||
| 102 | PLK1 | 9590 5266 | 9537 | 0.005 | 0.3124 | No | ||
| 103 | POLA1 | 24112 | 9841 | 0.001 | 0.2960 | No | ||
| 104 | CDC45L | 22642 1752 | 9951 | -0.001 | 0.2901 | No | ||
| 105 | CDK7 | 21365 | 9956 | -0.001 | 0.2899 | No | ||
| 106 | POLE2 | 21053 | 10899 | -0.013 | 0.2389 | No | ||
| 107 | PRKAR2B | 5288 2107 | 11006 | -0.014 | 0.2332 | No | ||
| 108 | UBE2D1 | 19738 | 12101 | -0.032 | 0.1741 | No | ||
| 109 | PSMA1 | 1627 17669 | 12144 | -0.033 | 0.1719 | No | ||
| 110 | PRKACA | 18549 3844 | 12270 | -0.036 | 0.1653 | No | ||
| 111 | KIF2A | 9217 11438 | 13195 | -0.061 | 0.1155 | No | ||
| 112 | CENPC1 | 8737 | 13390 | -0.068 | 0.1052 | No | ||
| 113 | PSMD11 | 12772 7600 | 14271 | -0.126 | 0.0579 | No | ||
| 114 | CLASP2 | 13338 | 14336 | -0.134 | 0.0549 | No | ||
| 115 | ORC2L | 385 13949 | 14740 | -0.190 | 0.0337 | No | ||
| 116 | PPP1CC | 9609 5283 | 14815 | -0.203 | 0.0303 | No | ||
| 117 | SMC1A | 2572 24225 6279 10780 | 15233 | -0.298 | 0.0087 | No | ||
| 118 | CDC25B | 14841 | 15365 | -0.339 | 0.0027 | No | ||
| 119 | BUB3 | 18045 | 15395 | -0.349 | 0.0023 | No | ||
| 120 | MAD2L1 | 17422 | 15496 | -0.394 | -0.0019 | No | ||
| 121 | NUMA1 | 18167 | 15573 | -0.432 | -0.0046 | No | ||
| 122 | CLASP1 | 14159 | 15606 | -0.444 | -0.0049 | No | ||
| 123 | SSNA1 | 4 | 15690 | -0.486 | -0.0079 | No | ||
| 124 | ORC1L | 327 16144 | 15837 | -0.579 | -0.0139 | No | ||
| 125 | STAG2 | 5521 | 16122 | -0.850 | -0.0266 | No | ||
| 126 | UBE2C | 12714 | 16124 | -0.853 | -0.0239 | No | ||
| 127 | TYMS | 5810 5809 3606 3598 | 16166 | -0.895 | -0.0233 | No | ||
| 128 | CETN2 | 24138 | 16203 | -0.930 | -0.0222 | No | ||
| 129 | YWHAG | 16339 | 16210 | -0.937 | -0.0196 | No | ||
| 130 | LIG1 | 18388 1749 1493 | 16393 | -1.092 | -0.0259 | No | ||
| 131 | CDC20 | 8421 8422 4227 | 16439 | -1.139 | -0.0247 | No | ||
| 132 | PTTG1 | 20490 | 16751 | -1.589 | -0.0365 | No | ||
| 133 | SGOL2 | 14247 | 16932 | -1.812 | -0.0404 | No | ||
| 134 | MAPRE1 | 4652 | 16998 | -1.902 | -0.0379 | No | ||
| 135 | CSNK1E | 6570 2211 11332 | 17205 | -2.253 | -0.0418 | No | ||
| 136 | PSMB10 | 5299 18761 | 17214 | -2.264 | -0.0350 | No | ||
| 137 | POLD1 | 17847 | 17439 | -2.709 | -0.0385 | No | ||
| 138 | CENPA | 8736 | 17479 | -2.823 | -0.0315 | No | ||
| 139 | RB1 | 21754 | 17554 | -2.982 | -0.0260 | No | ||
| 140 | ACTR1A | 23653 | 17730 | -3.449 | -0.0244 | No | ||
| 141 | CDKN1A | 4511 8729 | 18269 | -5.369 | -0.0364 | No | ||
| 142 | TK2 | 18779 | 18514 | -7.500 | -0.0256 | No | ||
| 143 | PSMB2 | 2324 16078 | 18611 | -9.723 | 0.0003 | No |