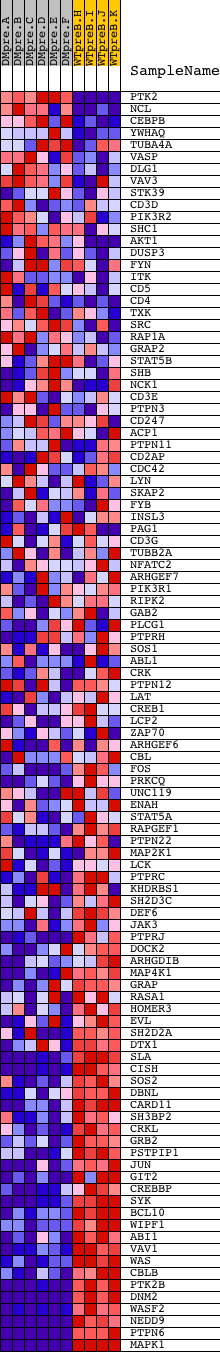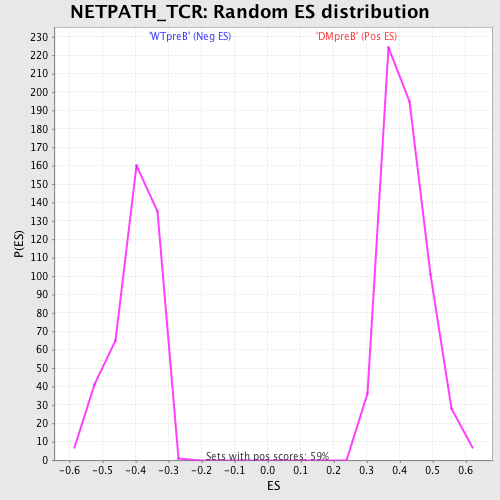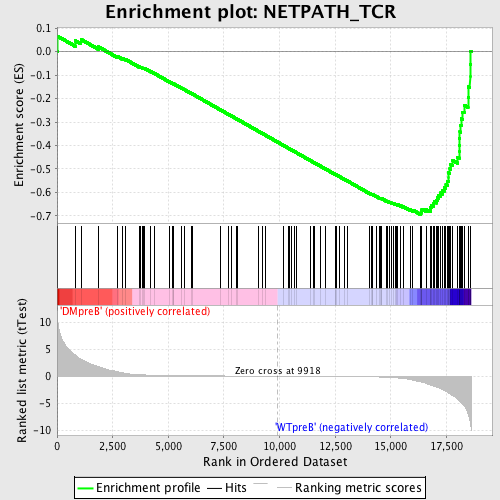
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | NETPATH_TCR |
| Enrichment Score (ES) | -0.6946163 |
| Normalized Enrichment Score (NES) | -1.7291919 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.008956725 |
| FWER p-Value | 0.059 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PTK2 | 22271 | 13 | 10.897 | 0.0665 | No | ||
| 2 | NCL | 5153 13899 | 818 | 3.882 | 0.0470 | No | ||
| 3 | CEBPB | 8733 | 1073 | 3.127 | 0.0526 | No | ||
| 4 | YWHAQ | 10369 | 1863 | 1.733 | 0.0206 | No | ||
| 5 | TUBA4A | 10232 | 2723 | 0.792 | -0.0209 | No | ||
| 6 | VASP | 5847 | 2944 | 0.606 | -0.0290 | No | ||
| 7 | DLG1 | 22793 1672 | 3069 | 0.517 | -0.0325 | No | ||
| 8 | VAV3 | 1774 1848 15443 | 3695 | 0.275 | -0.0646 | No | ||
| 9 | STK39 | 14570 | 3741 | 0.263 | -0.0654 | No | ||
| 10 | CD3D | 19473 | 3849 | 0.241 | -0.0697 | No | ||
| 11 | PIK3R2 | 18850 | 3888 | 0.232 | -0.0703 | No | ||
| 12 | SHC1 | 9813 9812 5430 | 3936 | 0.225 | -0.0715 | No | ||
| 13 | AKT1 | 8568 | 4218 | 0.183 | -0.0855 | No | ||
| 14 | DUSP3 | 1358 13022 | 4390 | 0.161 | -0.0938 | No | ||
| 15 | FYN | 3375 3395 20052 | 5072 | 0.109 | -0.1299 | No | ||
| 16 | ITK | 4934 | 5197 | 0.102 | -0.1360 | No | ||
| 17 | CD5 | 23741 | 5231 | 0.100 | -0.1371 | No | ||
| 18 | CD4 | 16999 | 5252 | 0.099 | -0.1376 | No | ||
| 19 | TXK | 16513 | 5591 | 0.082 | -0.1554 | No | ||
| 20 | SRC | 5507 | 5727 | 0.076 | -0.1622 | No | ||
| 21 | RAP1A | 8467 | 6027 | 0.066 | -0.1779 | No | ||
| 22 | GRAP2 | 5113 9398 | 6102 | 0.063 | -0.1815 | No | ||
| 23 | STAT5B | 20222 | 7325 | 0.036 | -0.2473 | No | ||
| 24 | SHB | 10493 | 7714 | 0.029 | -0.2681 | No | ||
| 25 | NCK1 | 9447 5152 | 7819 | 0.028 | -0.2736 | No | ||
| 26 | CD3E | 8714 | 8077 | 0.024 | -0.2873 | No | ||
| 27 | PTPN3 | 9661 | 8116 | 0.023 | -0.2892 | No | ||
| 28 | CD247 | 4498 8715 | 9036 | 0.011 | -0.3388 | No | ||
| 29 | ACP1 | 4317 | 9230 | 0.009 | -0.3491 | No | ||
| 30 | PTPN11 | 5326 16391 9660 | 9360 | 0.007 | -0.3561 | No | ||
| 31 | CD2AP | 22975 | 10173 | -0.003 | -0.3999 | No | ||
| 32 | CDC42 | 4503 8722 4504 2465 | 10407 | -0.006 | -0.4125 | No | ||
| 33 | LYN | 16281 | 10463 | -0.007 | -0.4154 | No | ||
| 34 | SKAP2 | 17153 | 10553 | -0.008 | -0.4201 | No | ||
| 35 | FYB | 10717 | 10678 | -0.010 | -0.4268 | No | ||
| 36 | INSL3 | 9181 | 10755 | -0.011 | -0.4308 | No | ||
| 37 | PAG1 | 15376 | 11377 | -0.020 | -0.4643 | No | ||
| 38 | CD3G | 19139 | 11522 | -0.022 | -0.4719 | No | ||
| 39 | TUBB2A | 21493 | 11571 | -0.023 | -0.4743 | No | ||
| 40 | NFATC2 | 5168 2866 | 11816 | -0.027 | -0.4874 | No | ||
| 41 | ARHGEF7 | 3823 | 12085 | -0.032 | -0.5016 | No | ||
| 42 | PIK3R1 | 3170 | 12502 | -0.041 | -0.5239 | No | ||
| 43 | RIPK2 | 2528 15935 | 12517 | -0.042 | -0.5244 | No | ||
| 44 | GAB2 | 1821 18184 2025 | 12548 | -0.043 | -0.5257 | No | ||
| 45 | PLCG1 | 14753 | 12675 | -0.046 | -0.5322 | No | ||
| 46 | PTPRH | 11861 | 12914 | -0.052 | -0.5448 | No | ||
| 47 | SOS1 | 5476 | 13046 | -0.056 | -0.5515 | No | ||
| 48 | ABL1 | 2693 4301 2794 | 14050 | -0.106 | -0.6050 | No | ||
| 49 | CRK | 4559 1249 | 14137 | -0.112 | -0.6090 | No | ||
| 50 | PTPN12 | 16597 3502 3586 3665 | 14167 | -0.115 | -0.6098 | No | ||
| 51 | LAT | 17643 | 14176 | -0.117 | -0.6096 | No | ||
| 52 | CREB1 | 3990 8782 4558 4093 | 14365 | -0.137 | -0.6189 | No | ||
| 53 | LCP2 | 4988 9268 | 14508 | -0.152 | -0.6256 | No | ||
| 54 | ZAP70 | 14271 4042 | 14538 | -0.156 | -0.6262 | No | ||
| 55 | ARHGEF6 | 13297 13104 | 14561 | -0.159 | -0.6264 | No | ||
| 56 | CBL | 19154 | 14602 | -0.166 | -0.6276 | No | ||
| 57 | FOS | 21202 | 14800 | -0.201 | -0.6370 | No | ||
| 58 | PRKCQ | 2873 2831 | 14829 | -0.205 | -0.6372 | No | ||
| 59 | UNC119 | 20758 | 14959 | -0.230 | -0.6428 | No | ||
| 60 | ENAH | 4665 8901 | 15045 | -0.247 | -0.6458 | No | ||
| 61 | STAT5A | 20664 | 15113 | -0.268 | -0.6478 | No | ||
| 62 | RAPGEF1 | 4218 2860 | 15230 | -0.297 | -0.6522 | No | ||
| 63 | PTPN22 | 1775 15470 | 15251 | -0.303 | -0.6514 | No | ||
| 64 | MAP2K1 | 19082 | 15318 | -0.325 | -0.6530 | No | ||
| 65 | LCK | 15746 | 15440 | -0.368 | -0.6573 | No | ||
| 66 | PTPRC | 5327 9662 | 15564 | -0.428 | -0.6613 | No | ||
| 67 | KHDRBS1 | 5405 9778 | 15864 | -0.607 | -0.6737 | No | ||
| 68 | SH2D3C | 2864 | 15995 | -0.713 | -0.6763 | No | ||
| 69 | DEF6 | 23319 | 16335 | -1.029 | -0.6883 | Yes | ||
| 70 | JAK3 | 9198 4936 | 16361 | -1.051 | -0.6831 | Yes | ||
| 71 | PTPRJ | 9664 | 16387 | -1.089 | -0.6778 | Yes | ||
| 72 | DOCK2 | 8243 | 16397 | -1.094 | -0.6715 | Yes | ||
| 73 | ARHGDIB | 16950 | 16594 | -1.343 | -0.6738 | Yes | ||
| 74 | MAP4K1 | 18313 | 16775 | -1.624 | -0.6735 | Yes | ||
| 75 | GRAP | 20851 | 16790 | -1.654 | -0.6641 | Yes | ||
| 76 | RASA1 | 10174 | 16815 | -1.689 | -0.6550 | Yes | ||
| 77 | HOMER3 | 18590 | 16906 | -1.781 | -0.6488 | Yes | ||
| 78 | EVL | 21156 | 16940 | -1.821 | -0.6394 | Yes | ||
| 79 | SH2D2A | 6569 | 17060 | -1.993 | -0.6335 | Yes | ||
| 80 | DTX1 | 16396 | 17100 | -2.068 | -0.6229 | Yes | ||
| 81 | SLA | 5449 | 17149 | -2.154 | -0.6122 | Yes | ||
| 82 | CISH | 8743 | 17238 | -2.298 | -0.6028 | Yes | ||
| 83 | SOS2 | 21049 | 17328 | -2.499 | -0.5922 | Yes | ||
| 84 | DBNL | 20954 | 17397 | -2.642 | -0.5796 | Yes | ||
| 85 | CARD11 | 8436 | 17478 | -2.819 | -0.5665 | Yes | ||
| 86 | SH3BP2 | 16874 | 17547 | -2.956 | -0.5519 | Yes | ||
| 87 | CRKL | 4560 | 17588 | -3.053 | -0.5353 | Yes | ||
| 88 | GRB2 | 20149 | 17590 | -3.055 | -0.5165 | Yes | ||
| 89 | PSTPIP1 | 19437 | 17655 | -3.243 | -0.4999 | Yes | ||
| 90 | JUN | 15832 | 17687 | -3.335 | -0.4810 | Yes | ||
| 91 | GIT2 | 11174 6462 | 17770 | -3.542 | -0.4636 | Yes | ||
| 92 | CREBBP | 22682 8783 | 17993 | -4.203 | -0.4497 | Yes | ||
| 93 | SYK | 21636 | 18074 | -4.548 | -0.4260 | Yes | ||
| 94 | BCL10 | 15397 | 18081 | -4.569 | -0.3981 | Yes | ||
| 95 | WIPF1 | 14560 | 18090 | -4.598 | -0.3702 | Yes | ||
| 96 | ABI1 | 4300 | 18105 | -4.642 | -0.3423 | Yes | ||
| 97 | VAV1 | 23173 | 18136 | -4.751 | -0.3146 | Yes | ||
| 98 | WAS | 24193 | 18166 | -4.862 | -0.2862 | Yes | ||
| 99 | CBLB | 5531 22734 | 18225 | -5.154 | -0.2576 | Yes | ||
| 100 | PTK2B | 21776 | 18298 | -5.513 | -0.2275 | Yes | ||
| 101 | DNM2 | 4635 3103 | 18471 | -6.991 | -0.1936 | Yes | ||
| 102 | WASF2 | 6326 | 18486 | -7.185 | -0.1501 | Yes | ||
| 103 | NEDD9 | 3206 21483 | 18558 | -8.130 | -0.1038 | Yes | ||
| 104 | PTPN6 | 17002 | 18561 | -8.216 | -0.0532 | Yes | ||
| 105 | MAPK1 | 1642 11167 | 18592 | -9.104 | 0.0013 | Yes |