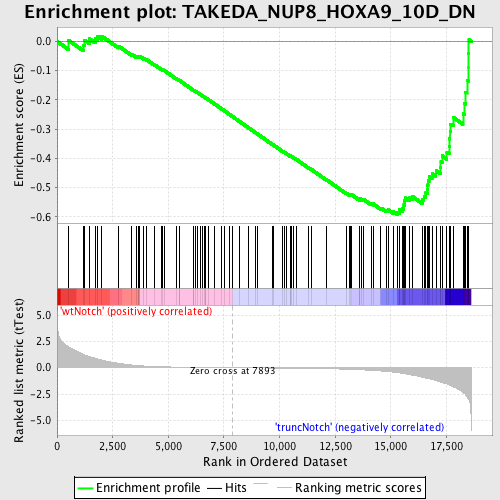
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch_repos |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#wtNotch_versus_truncNotch_repos |
| Upregulated in class | truncNotch |
| GeneSet | TAKEDA_NUP8_HOXA9_10D_DN |
| Enrichment Score (ES) | -0.5915197 |
| Normalized Enrichment Score (NES) | -1.6320069 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.14823912 |
| FWER p-Value | 0.908 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TNFAIP8 | 2760551 | 504 | 1.986 | 0.0043 | No | ||
| 2 | EMP1 | 4120438 | 1189 | 1.248 | -0.0128 | No | ||
| 3 | GGTA1 | 1570292 | 1232 | 1.218 | 0.0042 | No | ||
| 4 | GPNMB | 1780138 2340176 3390403 4560156 | 1449 | 1.040 | 0.0090 | No | ||
| 5 | CTSB | 1400524 2360064 | 1717 | 0.895 | 0.0088 | No | ||
| 6 | TSPAN2 | 3940161 | 1823 | 0.830 | 0.0163 | No | ||
| 7 | RPS6KA2 | 2970161 4730278 | 2004 | 0.726 | 0.0181 | No | ||
| 8 | SLC7A8 | 380136 5690672 | 2757 | 0.422 | -0.0158 | No | ||
| 9 | IL7R | 2060088 6380500 | 3351 | 0.254 | -0.0438 | No | ||
| 10 | RASGRP1 | 1050148 4050632 6510671 | 3571 | 0.208 | -0.0523 | No | ||
| 11 | MMP12 | 4920070 | 3649 | 0.198 | -0.0533 | No | ||
| 12 | GPD2 | 2350075 2690411 | 3658 | 0.196 | -0.0507 | No | ||
| 13 | RGL1 | 1500097 6900450 | 3708 | 0.188 | -0.0503 | No | ||
| 14 | TLR7 | 3140300 | 3902 | 0.158 | -0.0583 | No | ||
| 15 | LYPLAL1 | 5290717 5670048 | 4036 | 0.143 | -0.0632 | No | ||
| 16 | RGR | 730156 6400368 | 4397 | 0.110 | -0.0809 | No | ||
| 17 | PDK4 | 6400300 | 4679 | 0.090 | -0.0946 | No | ||
| 18 | SLFN13 | 2510068 | 4749 | 0.086 | -0.0970 | No | ||
| 19 | PLA2G7 | 4730092 | 4807 | 0.083 | -0.0988 | No | ||
| 20 | CD24 | 1780091 | 5372 | 0.057 | -0.1283 | No | ||
| 21 | MAOA | 1410039 4610324 | 5478 | 0.053 | -0.1331 | No | ||
| 22 | OLIG1 | 1660594 | 5513 | 0.052 | -0.1342 | No | ||
| 23 | RARRES1 | 770139 5860390 | 6131 | 0.033 | -0.1670 | No | ||
| 24 | NPL | 3520025 | 6150 | 0.033 | -0.1674 | No | ||
| 25 | RTN1 | 4150452 4850176 | 6223 | 0.031 | -0.1708 | No | ||
| 26 | NRP2 | 4070400 5860041 6650446 | 6307 | 0.030 | -0.1748 | No | ||
| 27 | LGALS12 | 3940242 | 6452 | 0.026 | -0.1822 | No | ||
| 28 | RNASE1 | 6550364 | 6530 | 0.025 | -0.1859 | No | ||
| 29 | PTGER3 | 2900739 6770750 | 6631 | 0.022 | -0.1910 | No | ||
| 30 | S100B | 6520088 | 6680 | 0.021 | -0.1932 | No | ||
| 31 | FCER1A | 1050671 | 6687 | 0.021 | -0.1932 | No | ||
| 32 | PCSK5 | 5720600 | 6816 | 0.019 | -0.1998 | No | ||
| 33 | ATP6V0D2 | 7050541 | 7086 | 0.013 | -0.2142 | No | ||
| 34 | DACH1 | 2450593 5700592 | 7395 | 0.008 | -0.2307 | No | ||
| 35 | SLC25A27 | 6200707 6940019 | 7520 | 0.006 | -0.2373 | No | ||
| 36 | C5AR1 | 4540402 | 7746 | 0.002 | -0.2494 | No | ||
| 37 | A2M | 6620039 | 7900 | -0.000 | -0.2576 | No | ||
| 38 | TRPA1 | 2680301 | 8184 | -0.005 | -0.2729 | No | ||
| 39 | CHIT1 | 7050347 | 8624 | -0.012 | -0.2964 | No | ||
| 40 | SEPP1 | 450273 | 8914 | -0.017 | -0.3117 | No | ||
| 41 | ST14 | 5360692 6380402 | 9008 | -0.018 | -0.3165 | No | ||
| 42 | RAP1A | 1090025 | 9662 | -0.029 | -0.3513 | No | ||
| 43 | ACSL6 | 2850008 6040279 | 9728 | -0.031 | -0.3543 | No | ||
| 44 | HRH4 | 4010082 | 10130 | -0.039 | -0.3753 | No | ||
| 45 | SRPX | 580717 1980162 | 10220 | -0.040 | -0.3795 | No | ||
| 46 | MS4A2 | 2940594 | 10315 | -0.042 | -0.3839 | No | ||
| 47 | IL1R2 | 1410600 | 10469 | -0.046 | -0.3914 | No | ||
| 48 | TEX15 | 380358 | 10479 | -0.046 | -0.3912 | No | ||
| 49 | FUCA1 | 610075 | 10520 | -0.047 | -0.3926 | No | ||
| 50 | CXCL5 | 6370333 | 10633 | -0.050 | -0.3979 | No | ||
| 51 | KCNJ15 | 70176 6900750 | 10756 | -0.053 | -0.4036 | No | ||
| 52 | ABP1 | 1980609 60056 | 11303 | -0.067 | -0.4321 | No | ||
| 53 | OTOA | 2120040 2680528 | 11442 | -0.071 | -0.4384 | No | ||
| 54 | CLEC5A | 6420139 | 12100 | -0.095 | -0.4724 | No | ||
| 55 | HS3ST1 | 1850193 | 13008 | -0.143 | -0.5191 | No | ||
| 56 | IL1R1 | 3850377 6180128 | 13133 | -0.152 | -0.5234 | No | ||
| 57 | PPAP2B | 3190397 4730280 | 13206 | -0.156 | -0.5248 | No | ||
| 58 | OLFML2B | 4060056 | 13246 | -0.160 | -0.5244 | No | ||
| 59 | DISC1 | 3940487 840463 5130082 | 13602 | -0.190 | -0.5405 | No | ||
| 60 | PPBP | 5130446 | 13605 | -0.191 | -0.5376 | No | ||
| 61 | DOCK4 | 5910102 | 13691 | -0.199 | -0.5391 | No | ||
| 62 | CXCL2 | 610398 | 13750 | -0.205 | -0.5389 | No | ||
| 63 | FABP3 | 6860452 | 14124 | -0.245 | -0.5552 | No | ||
| 64 | ANXA3 | 5570494 | 14200 | -0.257 | -0.5552 | No | ||
| 65 | SLAMF8 | 1230048 | 14557 | -0.310 | -0.5695 | No | ||
| 66 | CXCL1 | 2690537 | 14790 | -0.349 | -0.5765 | No | ||
| 67 | STAB1 | 5390707 | 14874 | -0.364 | -0.5752 | No | ||
| 68 | GPR97 | 6100100 | 15102 | -0.413 | -0.5809 | No | ||
| 69 | RASSF4 | 510372 1660673 6290040 | 15300 | -0.460 | -0.5842 | Yes | ||
| 70 | MMP9 | 580338 | 15380 | -0.483 | -0.5808 | Yes | ||
| 71 | ACP5 | 2230717 | 15383 | -0.483 | -0.5733 | Yes | ||
| 72 | TACSTD2 | 6660341 | 15518 | -0.529 | -0.5721 | Yes | ||
| 73 | DAB2 | 60309 | 15583 | -0.551 | -0.5668 | Yes | ||
| 74 | MAPK13 | 6760215 | 15589 | -0.553 | -0.5583 | Yes | ||
| 75 | INADL | 380092 2680278 3360440 4590528 4640537 | 15617 | -0.566 | -0.5508 | Yes | ||
| 76 | RAPH1 | 6760411 | 15627 | -0.568 | -0.5422 | Yes | ||
| 77 | FJX1 | 3390035 | 15664 | -0.583 | -0.5349 | Yes | ||
| 78 | FRMD4A | 2190176 | 15853 | -0.644 | -0.5349 | Yes | ||
| 79 | SGK | 1400131 2480056 | 15969 | -0.698 | -0.5300 | Yes | ||
| 80 | HEXA | 2630358 | 16426 | -0.905 | -0.5403 | Yes | ||
| 81 | PPP1R1B | 5290403 5550324 | 16491 | -0.935 | -0.5289 | Yes | ||
| 82 | CORO2A | 3840605 6620390 | 16575 | -0.976 | -0.5179 | Yes | ||
| 83 | PADI4 | 1740075 4810441 | 16640 | -0.999 | -0.5055 | Yes | ||
| 84 | CD9 | 4730041 | 16660 | -1.005 | -0.4906 | Yes | ||
| 85 | BLNK | 70288 6590092 | 16674 | -1.009 | -0.4753 | Yes | ||
| 86 | TREM2 | 6290358 | 16719 | -1.029 | -0.4613 | Yes | ||
| 87 | SLPI | 2120446 | 16855 | -1.098 | -0.4512 | Yes | ||
| 88 | C1QB | 5910292 | 17030 | -1.214 | -0.4413 | Yes | ||
| 89 | CCL4 | 50368 430047 | 17242 | -1.353 | -0.4313 | Yes | ||
| 90 | CHI3L1 | 1170039 | 17254 | -1.360 | -0.4103 | Yes | ||
| 91 | SLCO2B1 | 3450019 | 17306 | -1.391 | -0.3910 | Yes | ||
| 92 | CCL7 | 2650519 | 17523 | -1.539 | -0.3782 | Yes | ||
| 93 | SERPINF1 | 7040367 | 17622 | -1.624 | -0.3577 | Yes | ||
| 94 | RIT1 | 5390338 | 17639 | -1.639 | -0.3326 | Yes | ||
| 95 | ACE | 1780161 4920315 | 17659 | -1.659 | -0.3073 | Yes | ||
| 96 | C1QA | 5390687 | 17695 | -1.694 | -0.2823 | Yes | ||
| 97 | C1QC | 5700131 | 17801 | -1.798 | -0.2594 | Yes | ||
| 98 | LHFPL2 | 450020 | 18245 | -2.327 | -0.2464 | Yes | ||
| 99 | LGMN | 3610301 | 18316 | -2.444 | -0.2114 | Yes | ||
| 100 | APOE | 4200671 | 18370 | -2.578 | -0.1734 | Yes | ||
| 101 | TGFBI | 2060446 6900112 | 18425 | -2.742 | -0.1328 | Yes | ||
| 102 | PADI2 | 2940092 6420136 | 18499 | -2.975 | -0.0895 | Yes | ||
| 103 | CST7 | 380369 | 18501 | -2.985 | -0.0422 | Yes | ||
| 104 | KLF2 | 6860270 | 18512 | -3.049 | 0.0056 | Yes |

