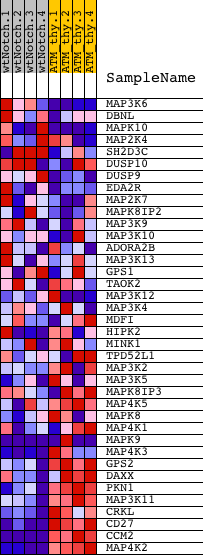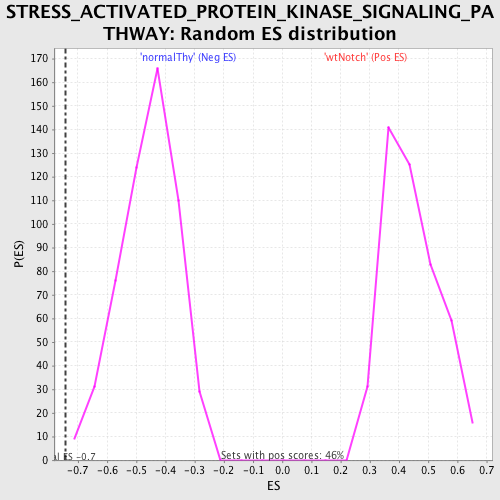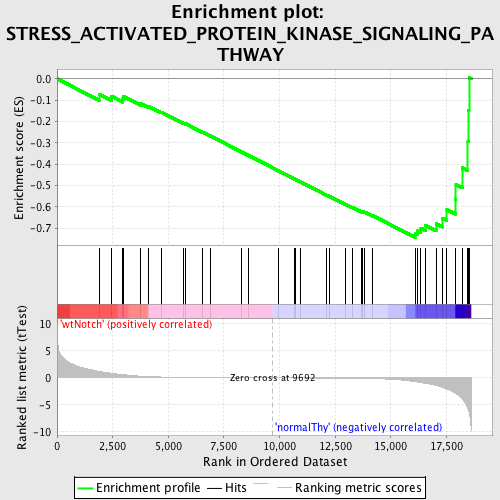
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | STRESS_ACTIVATED_PROTEIN_KINASE_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.7440113 |
| Normalized Enrichment Score (NES) | -1.6276935 |
| Nominal p-value | 0.0018348624 |
| FDR q-value | 0.1975346 |
| FWER p-Value | 0.629 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MAP3K6 | 6400390 | 1886 | 1.198 | -0.0724 | No | ||
| 2 | DBNL | 7100088 | 2437 | 0.858 | -0.0812 | No | ||
| 3 | MAPK10 | 6110193 | 2956 | 0.591 | -0.0948 | No | ||
| 4 | MAP2K4 | 5130133 | 2991 | 0.578 | -0.0826 | No | ||
| 5 | SH2D3C | 3060014 | 3756 | 0.306 | -0.1163 | No | ||
| 6 | DUSP10 | 2850673 3360064 | 4094 | 0.235 | -0.1287 | No | ||
| 7 | DUSP9 | 630605 | 4680 | 0.161 | -0.1563 | No | ||
| 8 | EDA2R | 870397 | 5695 | 0.092 | -0.2087 | No | ||
| 9 | MAP2K7 | 2260086 | 5781 | 0.087 | -0.2111 | No | ||
| 10 | MAPK8IP2 | 630373 | 6535 | 0.060 | -0.2502 | No | ||
| 11 | MAP3K9 | 3290707 | 6885 | 0.050 | -0.2678 | No | ||
| 12 | MAP3K10 | 3360168 4850180 | 8267 | 0.023 | -0.3416 | No | ||
| 13 | ADORA2B | 6860253 | 8596 | 0.017 | -0.3588 | No | ||
| 14 | MAP3K13 | 3190017 | 9942 | -0.003 | -0.4311 | No | ||
| 15 | GPS1 | 2470037 4760315 6200528 7100040 | 10663 | -0.014 | -0.4695 | No | ||
| 16 | TAOK2 | 110575 6380692 | 10719 | -0.015 | -0.4721 | No | ||
| 17 | MAP3K12 | 2470373 6420162 | 10936 | -0.019 | -0.4833 | No | ||
| 18 | MAP3K4 | 5720041 | 12130 | -0.041 | -0.5465 | No | ||
| 19 | MDFI | 6200041 | 12246 | -0.044 | -0.5516 | No | ||
| 20 | HIPK2 | 6350647 | 12971 | -0.065 | -0.5890 | No | ||
| 21 | MINK1 | 5860647 | 13258 | -0.076 | -0.6026 | No | ||
| 22 | TPD52L1 | 4590711 5570575 | 13674 | -0.098 | -0.6225 | No | ||
| 23 | MAP3K2 | 380270 | 13712 | -0.100 | -0.6221 | No | ||
| 24 | MAP3K5 | 6020041 6380162 | 13822 | -0.108 | -0.6253 | No | ||
| 25 | MAPK8IP3 | 1660112 2060427 | 14176 | -0.136 | -0.6410 | No | ||
| 26 | MAP4K5 | 3190286 4760300 5270070 5550494 | 16090 | -0.707 | -0.7269 | Yes | ||
| 27 | MAPK8 | 2640195 | 16187 | -0.761 | -0.7136 | Yes | ||
| 28 | MAP4K1 | 4060692 | 16352 | -0.869 | -0.7013 | Yes | ||
| 29 | MAPK9 | 2060273 3780209 4070397 | 16570 | -1.002 | -0.6887 | Yes | ||
| 30 | MAP4K3 | 5390551 | 17033 | -1.381 | -0.6801 | Yes | ||
| 31 | GPS2 | 5910672 | 17312 | -1.730 | -0.6531 | Yes | ||
| 32 | DAXX | 4010647 | 17522 | -2.068 | -0.6142 | Yes | ||
| 33 | PKN1 | 2640605 2760647 | 17895 | -2.835 | -0.5655 | Yes | ||
| 34 | MAP3K11 | 7000039 | 17928 | -2.933 | -0.4960 | Yes | ||
| 35 | CRKL | 4050427 | 18209 | -3.866 | -0.4173 | Yes | ||
| 36 | CD27 | 610020 | 18467 | -5.721 | -0.2924 | Yes | ||
| 37 | CCM2 | 5670465 | 18496 | -6.023 | -0.1478 | Yes | ||
| 38 | MAP4K2 | 4200037 | 18514 | -6.359 | 0.0055 | Yes |