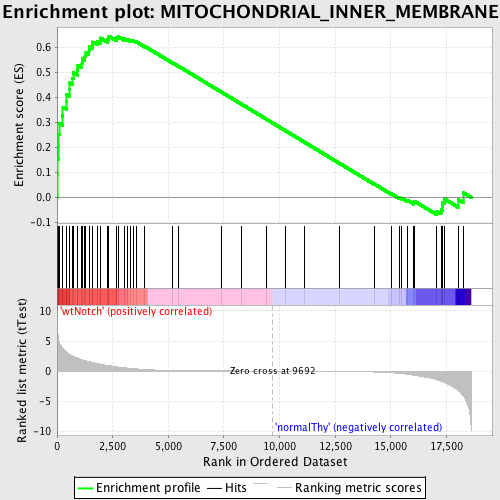
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | wtNotch |
| GeneSet | MITOCHONDRIAL_INNER_MEMBRANE |
| Enrichment Score (ES) | 0.64552426 |
| Normalized Enrichment Score (NES) | 1.5769852 |
| Nominal p-value | 0.0046296297 |
| FDR q-value | 0.1449009 |
| FWER p-Value | 0.942 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TIMM10 | 610070 1450019 | 3 | 9.579 | 0.0923 | Yes | ||
| 2 | SLC25A11 | 130097 770685 | 32 | 6.620 | 0.1548 | Yes | ||
| 3 | TIMM13 | 2450601 | 67 | 5.283 | 0.2039 | Yes | ||
| 4 | TIMM50 | 1500139 | 73 | 5.157 | 0.2535 | Yes | ||
| 5 | PHB2 | 1230050 | 105 | 4.698 | 0.2972 | Yes | ||
| 6 | ATP5G1 | 5290019 | 245 | 3.817 | 0.3265 | Yes | ||
| 7 | SLC25A3 | 670112 | 262 | 3.748 | 0.3619 | Yes | ||
| 8 | UQCRC1 | 520053 | 432 | 3.119 | 0.3829 | Yes | ||
| 9 | BCS1L | 5360102 | 439 | 3.094 | 0.4124 | Yes | ||
| 10 | ATP5G2 | 840068 | 570 | 2.750 | 0.4320 | Yes | ||
| 11 | SLC25A1 | 4050402 | 574 | 2.736 | 0.4582 | Yes | ||
| 12 | COX15 | 2360402 | 678 | 2.568 | 0.4775 | Yes | ||
| 13 | NDUFV1 | 5420369 | 722 | 2.493 | 0.4992 | Yes | ||
| 14 | ABCB7 | 4150324 | 906 | 2.165 | 0.5103 | Yes | ||
| 15 | TIMM17A | 130092 | 934 | 2.131 | 0.5294 | Yes | ||
| 16 | ATP5E | 4120411 | 1117 | 1.888 | 0.5378 | Yes | ||
| 17 | NDUFA6 | 3990348 | 1132 | 1.877 | 0.5552 | Yes | ||
| 18 | UQCRB | 1450047 | 1250 | 1.758 | 0.5659 | Yes | ||
| 19 | SDHD | 2570128 | 1295 | 1.726 | 0.5802 | Yes | ||
| 20 | SURF1 | 5130048 | 1433 | 1.595 | 0.5882 | Yes | ||
| 21 | NDUFS7 | 1740142 | 1446 | 1.587 | 0.6029 | Yes | ||
| 22 | ATP5D | 6550167 | 1586 | 1.447 | 0.6093 | Yes | ||
| 23 | NDUFS2 | 4850020 6200402 | 1607 | 1.426 | 0.6220 | Yes | ||
| 24 | ATP5A1 | 1990722 | 1796 | 1.273 | 0.6242 | Yes | ||
| 25 | ATP5C1 | 5910601 | 1933 | 1.162 | 0.6281 | Yes | ||
| 26 | ATP5O | 7000398 | 1954 | 1.140 | 0.6380 | Yes | ||
| 27 | NDUFS8 | 4150100 | 2264 | 0.971 | 0.6307 | Yes | ||
| 28 | SLC25A15 | 4640368 5550064 | 2299 | 0.947 | 0.6380 | Yes | ||
| 29 | ATP5G3 | 4230551 | 2327 | 0.925 | 0.6455 | Yes | ||
| 30 | TIMM23 | 60288 7040133 | 2668 | 0.728 | 0.6342 | No | ||
| 31 | NDUFS3 | 1690136 5340528 | 2678 | 0.720 | 0.6407 | No | ||
| 32 | NDUFA1 | 6200411 | 2749 | 0.680 | 0.6435 | No | ||
| 33 | PMPCA | 4280132 6840408 | 3020 | 0.564 | 0.6344 | No | ||
| 34 | NDUFA9 | 2260280 | 3148 | 0.519 | 0.6326 | No | ||
| 35 | TIMM9 | 2450484 | 3293 | 0.460 | 0.6293 | No | ||
| 36 | COX6B2 | 1340010 4120039 | 3421 | 0.422 | 0.6265 | No | ||
| 37 | NDUFAB1 | 1980091 | 3559 | 0.372 | 0.6227 | No | ||
| 38 | OPA1 | 2030433 3520390 4730537 | 3947 | 0.265 | 0.6044 | No | ||
| 39 | UCP3 | 4200128 4210148 | 5189 | 0.119 | 0.5387 | No | ||
| 40 | HSD3B2 | 70068 770037 4120114 4230170 5690333 | 5464 | 0.104 | 0.5249 | No | ||
| 41 | ATP5J | 4210100 6900288 | 7383 | 0.039 | 0.4219 | No | ||
| 42 | HCCS | 670397 3170341 | 8297 | 0.022 | 0.3729 | No | ||
| 43 | HSD3B1 | 3060672 | 9427 | 0.004 | 0.3121 | No | ||
| 44 | SDHA | 7000056 | 10245 | -0.008 | 0.2682 | No | ||
| 45 | PHB | 2260739 | 11141 | -0.022 | 0.2202 | No | ||
| 46 | NDUFS1 | 2650253 | 12678 | -0.055 | 0.1379 | No | ||
| 47 | MRPL32 | 110041 | 14248 | -0.144 | 0.0548 | No | ||
| 48 | PPOX | 2640678 3850102 | 15014 | -0.261 | 0.0161 | No | ||
| 49 | NNT | 540253 1170471 5550092 6760397 | 15402 | -0.369 | -0.0012 | No | ||
| 50 | SLC25A22 | 2360050 | 15490 | -0.404 | -0.0020 | No | ||
| 51 | MPV17 | 1190133 4070427 4570577 | 15759 | -0.526 | -0.0114 | No | ||
| 52 | ATP5B | 3870138 | 16035 | -0.671 | -0.0197 | No | ||
| 53 | TIMM17B | 6040400 | 16066 | -0.690 | -0.0147 | No | ||
| 54 | TIMM8B | 1190364 | 17051 | -1.413 | -0.0541 | No | ||
| 55 | UQCRH | 4210463 | 17257 | -1.670 | -0.0490 | No | ||
| 56 | NDUFA13 | 6220021 | 17309 | -1.728 | -0.0350 | No | ||
| 57 | IMMT | 870450 4280438 | 17315 | -1.731 | -0.0186 | No | ||
| 58 | NDUFA2 | 6020100 | 17418 | -1.897 | -0.0058 | No | ||
| 59 | NDUFS4 | 510142 1240148 4760253 | 18022 | -3.137 | -0.0080 | No | ||
| 60 | ALAS2 | 6550176 | 18254 | -4.136 | 0.0195 | No |

