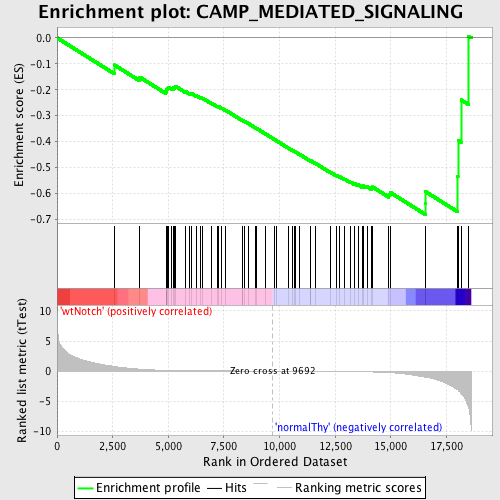
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | CAMP_MEDIATED_SIGNALING |
| Enrichment Score (ES) | -0.6808017 |
| Normalized Enrichment Score (NES) | -1.6242847 |
| Nominal p-value | 0.0034188034 |
| FDR q-value | 0.17698507 |
| FWER p-Value | 0.66 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | OPRK1 | 3780632 | 2574 | 0.774 | -0.1044 | No | ||
| 2 | LTB4R2 | 1990746 | 3707 | 0.322 | -0.1511 | No | ||
| 3 | EDNRB | 4280717 6400435 | 4894 | 0.140 | -0.2088 | No | ||
| 4 | GRK5 | 1940348 4670053 | 4902 | 0.140 | -0.2029 | No | ||
| 5 | ADCY7 | 6290520 7560739 | 4919 | 0.139 | -0.1976 | No | ||
| 6 | GALR3 | 4850113 | 4959 | 0.135 | -0.1938 | No | ||
| 7 | ADCYAP1 | 520086 | 4988 | 0.132 | -0.1894 | No | ||
| 8 | ACR | 2940017 3060050 | 5162 | 0.121 | -0.1934 | No | ||
| 9 | ADRA1D | 380035 540025 | 5238 | 0.116 | -0.1922 | No | ||
| 10 | GABBR1 | 2850519 2850735 | 5280 | 0.114 | -0.1894 | No | ||
| 11 | CCR2 | 1450519 | 5326 | 0.111 | -0.1869 | No | ||
| 12 | DRD5 | 2970133 | 5750 | 0.089 | -0.2057 | No | ||
| 13 | GNAI3 | 4670739 | 5965 | 0.080 | -0.2137 | No | ||
| 14 | GALR1 | 6020452 | 6020 | 0.078 | -0.2132 | No | ||
| 15 | CAP2 | 7100315 | 6261 | 0.069 | -0.2231 | No | ||
| 16 | FPR1 | 3800577 | 6436 | 0.063 | -0.2297 | No | ||
| 17 | PTGER4 | 6400670 | 6531 | 0.060 | -0.2321 | No | ||
| 18 | CCR3 | 50427 | 6932 | 0.049 | -0.2514 | No | ||
| 19 | ADRB1 | 6860292 | 7213 | 0.042 | -0.2646 | No | ||
| 20 | MC4R | 430333 | 7238 | 0.042 | -0.2641 | No | ||
| 21 | DRD1 | 430025 | 7409 | 0.039 | -0.2715 | No | ||
| 22 | GLP2R | 4150577 5290273 7050095 | 7573 | 0.035 | -0.2787 | No | ||
| 23 | GLP1R | 6420528 | 8317 | 0.022 | -0.3178 | No | ||
| 24 | EDNRA | 6900133 | 8319 | 0.022 | -0.3169 | No | ||
| 25 | ADRA2A | 5340520 | 8443 | 0.019 | -0.3227 | No | ||
| 26 | ADORA2B | 6860253 | 8596 | 0.017 | -0.3301 | No | ||
| 27 | GHRHR | 3130500 4010154 | 8910 | 0.012 | -0.3464 | No | ||
| 28 | GPR3 | 2970452 | 8962 | 0.011 | -0.3487 | No | ||
| 29 | GHRH | 4570575 | 9361 | 0.005 | -0.3699 | No | ||
| 30 | GRM4 | 6960092 | 9771 | -0.001 | -0.3919 | No | ||
| 31 | PTHLH | 5290739 | 9842 | -0.002 | -0.3956 | No | ||
| 32 | ADRA1B | 2100484 | 10414 | -0.010 | -0.4259 | No | ||
| 33 | ADORA3 | 630333 | 10583 | -0.013 | -0.4344 | No | ||
| 34 | CALCA | 5860167 | 10676 | -0.015 | -0.4387 | No | ||
| 35 | ADRB3 | 6900072 | 10718 | -0.015 | -0.4402 | No | ||
| 36 | GRM8 | 4780082 | 10889 | -0.018 | -0.4486 | No | ||
| 37 | CHRM5 | 5390333 | 11392 | -0.027 | -0.4745 | No | ||
| 38 | CHRM2 | 870750 | 11399 | -0.027 | -0.4736 | No | ||
| 39 | GCGR | 6620497 | 11625 | -0.031 | -0.4844 | No | ||
| 40 | CALCR | 1690494 | 12290 | -0.045 | -0.5182 | No | ||
| 41 | NF1 | 6980433 | 12559 | -0.052 | -0.5303 | No | ||
| 42 | GALR2 | 6450739 | 12694 | -0.056 | -0.5350 | No | ||
| 43 | NPY1R | 5890347 | 12915 | -0.063 | -0.5441 | No | ||
| 44 | MCHR1 | 1780022 | 13192 | -0.074 | -0.5557 | No | ||
| 45 | DRD2 | 5890369 | 13345 | -0.080 | -0.5603 | No | ||
| 46 | GRM3 | 4540242 | 13530 | -0.089 | -0.5663 | No | ||
| 47 | CRHR1 | 450161 3140070 5390647 | 13723 | -0.101 | -0.5721 | No | ||
| 48 | GRM7 | 4230110 | 13782 | -0.105 | -0.5706 | No | ||
| 49 | NPY2R | 6220612 | 13931 | -0.115 | -0.5735 | No | ||
| 50 | CRHR2 | 4590672 | 14144 | -0.134 | -0.5790 | No | ||
| 51 | CORT | 360008 2340092 | 14177 | -0.136 | -0.5747 | No | ||
| 52 | RGS1 | 4060347 4540181 | 14916 | -0.238 | -0.6039 | No | ||
| 53 | AVPR2 | 360064 | 14963 | -0.250 | -0.5953 | No | ||
| 54 | PRKACB | 4210170 | 16551 | -0.996 | -0.6366 | Yes | ||
| 55 | GNAS | 630441 1850373 4050152 | 16556 | -0.998 | -0.5926 | Yes | ||
| 56 | ADRB2 | 3290373 | 18009 | -3.105 | -0.5331 | Yes | ||
| 57 | NDUFS4 | 510142 1240148 4760253 | 18022 | -3.137 | -0.3947 | Yes | ||
| 58 | WASF2 | 4850592 | 18159 | -3.677 | -0.2390 | Yes | ||
| 59 | CAP1 | 2650278 | 18491 | -5.943 | 0.0067 | Yes |

