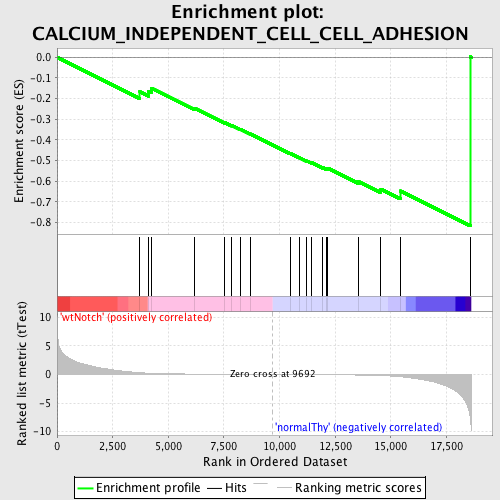
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | CALCIUM_INDEPENDENT_CELL_CELL_ADHESION |
| Enrichment Score (ES) | -0.81639713 |
| Normalized Enrichment Score (NES) | -1.5579156 |
| Nominal p-value | 0.0018450185 |
| FDR q-value | 0.2270049 |
| FWER p-Value | 0.982 |

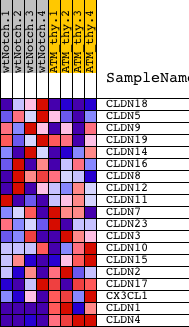
| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CLDN18 | 2030193 | 3718 | 0.319 | -0.1674 | No | ||
| 2 | CLDN5 | 6510717 | 4104 | 0.234 | -0.1643 | No | ||
| 3 | CLDN9 | 840373 | 4235 | 0.213 | -0.1496 | No | ||
| 4 | CLDN19 | 2690184 6370240 | 6192 | 0.071 | -0.2475 | No | ||
| 5 | CLDN14 | 5340292 | 7522 | 0.036 | -0.3153 | No | ||
| 6 | CLDN16 | 1850286 | 7849 | 0.030 | -0.3297 | No | ||
| 7 | CLDN8 | 1770113 2320500 | 8245 | 0.023 | -0.3486 | No | ||
| 8 | CLDN12 | 6350341 | 8694 | 0.015 | -0.3711 | No | ||
| 9 | CLDN11 | 670338 | 10483 | -0.011 | -0.4661 | No | ||
| 10 | CLDN7 | 1190301 | 10882 | -0.018 | -0.4857 | No | ||
| 11 | CLDN23 | 4670538 | 11226 | -0.023 | -0.5018 | No | ||
| 12 | CLDN3 | 2570524 | 11433 | -0.027 | -0.5101 | No | ||
| 13 | CLDN10 | 4070364 4590059 4920097 5080687 6620270 | 11928 | -0.037 | -0.5329 | No | ||
| 14 | CLDN15 | 4150270 4730592 | 12128 | -0.041 | -0.5394 | No | ||
| 15 | CLDN2 | 580053 | 12151 | -0.042 | -0.5363 | No | ||
| 16 | CLDN17 | 3710672 | 13547 | -0.090 | -0.6021 | No | ||
| 17 | CX3CL1 | 3990707 | 14550 | -0.173 | -0.6384 | No | ||
| 18 | CLDN1 | 5670746 | 15429 | -0.379 | -0.6470 | Yes | ||
| 19 | CLDN4 | 4920739 | 18581 | -8.039 | 0.0019 | Yes |