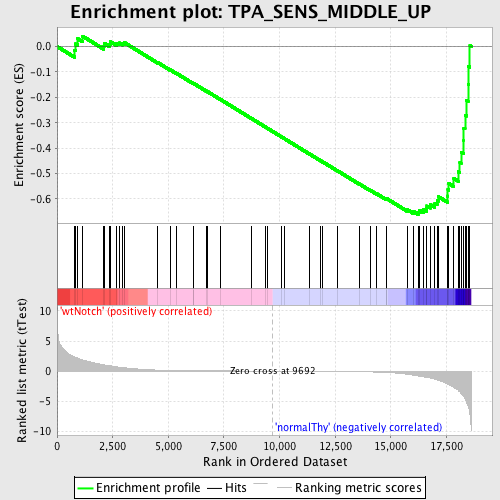
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | TPA_SENS_MIDDLE_UP |
| Enrichment Score (ES) | -0.66116095 |
| Normalized Enrichment Score (NES) | -1.5610195 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.29577976 |
| FWER p-Value | 0.994 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RPS6KA1 | 4120647 | 789 | 2.372 | -0.0145 | No | ||
| 2 | DPP4 | 5720369 | 835 | 2.287 | 0.0101 | No | ||
| 3 | CDC37 | 2570537 | 929 | 2.140 | 0.0303 | No | ||
| 4 | CCT5 | 1050670 | 1142 | 1.862 | 0.0409 | No | ||
| 5 | PLAUR | 5910280 | 2096 | 1.040 | 0.0018 | No | ||
| 6 | HSPA8 | 2120315 1050132 6550746 | 2111 | 1.030 | 0.0132 | No | ||
| 7 | ALAS1 | 6400440 | 2373 | 0.901 | 0.0098 | No | ||
| 8 | CCL7 | 2650519 | 2399 | 0.879 | 0.0188 | No | ||
| 9 | ALOX5AP | 3170440 | 2667 | 0.729 | 0.0130 | No | ||
| 10 | CCL3 | 2810092 | 2794 | 0.662 | 0.0140 | No | ||
| 11 | SNRPB | 1230097 2940086 3520671 | 2948 | 0.593 | 0.0128 | No | ||
| 12 | CYBA | 4760739 | 3011 | 0.567 | 0.0162 | No | ||
| 13 | TSC22D1 | 1340739 6040181 | 4529 | 0.176 | -0.0635 | No | ||
| 14 | CCL2 | 4760019 | 5083 | 0.126 | -0.0918 | No | ||
| 15 | HBEGF | 2030156 | 5378 | 0.109 | -0.1064 | No | ||
| 16 | IL1RAP | 50546 450711 1580397 3940301 5890181 6370373 | 6115 | 0.074 | -0.1452 | No | ||
| 17 | SERPINE1 | 4210403 | 6138 | 0.073 | -0.1455 | No | ||
| 18 | RARG | 6760136 | 6697 | 0.055 | -0.1749 | No | ||
| 19 | SPRY2 | 5860184 | 6779 | 0.053 | -0.1786 | No | ||
| 20 | P2RY5 | 3800136 | 7338 | 0.040 | -0.2082 | No | ||
| 21 | NR0B2 | 2470692 | 8755 | 0.014 | -0.2844 | No | ||
| 22 | VIM | 20431 | 9366 | 0.005 | -0.3172 | No | ||
| 23 | RHOH | 130184 | 9467 | 0.003 | -0.3225 | No | ||
| 24 | IL1RN | 2370333 | 10073 | -0.005 | -0.3551 | No | ||
| 25 | DCT | 1090347 3840494 | 10215 | -0.008 | -0.3626 | No | ||
| 26 | C3AR1 | 5720131 | 11335 | -0.025 | -0.4226 | No | ||
| 27 | SLC13A3 | 2850601 | 11859 | -0.036 | -0.4503 | No | ||
| 28 | AQP4 | 6370112 | 11950 | -0.037 | -0.4547 | No | ||
| 29 | AGER | 5860538 5910180 | 12590 | -0.053 | -0.4886 | No | ||
| 30 | AGA | 6350292 | 13582 | -0.091 | -0.5409 | No | ||
| 31 | TGFB2 | 4920292 | 14066 | -0.126 | -0.5654 | No | ||
| 32 | VIL1 | 7400129 | 14374 | -0.156 | -0.5801 | No | ||
| 33 | MYLK | 4010600 7000364 | 14789 | -0.211 | -0.5999 | No | ||
| 34 | MMP15 | 5900717 | 14823 | -0.219 | -0.5991 | No | ||
| 35 | SAT1 | 4570463 | 15729 | -0.511 | -0.6419 | No | ||
| 36 | PRKAR2B | 3130593 5220577 | 16009 | -0.656 | -0.6491 | No | ||
| 37 | TAPBP | 1980110 | 16233 | -0.793 | -0.6518 | Yes | ||
| 38 | EMP3 | 1500722 1570039 | 16298 | -0.836 | -0.6454 | Yes | ||
| 39 | DYNLL1 | 5570113 | 16457 | -0.935 | -0.6429 | Yes | ||
| 40 | PIK3CB | 3800600 | 16603 | -1.014 | -0.6387 | Yes | ||
| 41 | IFNGR2 | 1500288 2100176 | 16623 | -1.023 | -0.6277 | Yes | ||
| 42 | RAC2 | 1580541 | 16776 | -1.151 | -0.6223 | Yes | ||
| 43 | EXT1 | 4540603 | 16980 | -1.321 | -0.6176 | Yes | ||
| 44 | S100A11 | 2260064 | 17081 | -1.449 | -0.6059 | Yes | ||
| 45 | KCNN3 | 6520600 6420138 | 17130 | -1.518 | -0.5905 | Yes | ||
| 46 | DUSP2 | 2370184 | 17558 | -2.134 | -0.5884 | Yes | ||
| 47 | PCQAP | 3060064 6200273 | 17567 | -2.153 | -0.5634 | Yes | ||
| 48 | IMPDH1 | 3190735 7050546 | 17606 | -2.217 | -0.5392 | Yes | ||
| 49 | RFXAP | 3290181 | 17833 | -2.705 | -0.5195 | Yes | ||
| 50 | DUSP6 | 5910286 7100070 | 18031 | -3.173 | -0.4927 | Yes | ||
| 51 | PREP | 6220292 | 18105 | -3.446 | -0.4559 | Yes | ||
| 52 | NFKBIA | 1570152 | 18160 | -3.680 | -0.4154 | Yes | ||
| 53 | FLNA | 5390193 | 18277 | -4.252 | -0.3714 | Yes | ||
| 54 | TRAF1 | 3440735 | 18283 | -4.285 | -0.3211 | Yes | ||
| 55 | IL2RG | 4120273 | 18336 | -4.588 | -0.2698 | Yes | ||
| 56 | PLD3 | 510041 | 18412 | -5.170 | -0.2128 | Yes | ||
| 57 | EGR2 | 3800403 | 18475 | -5.788 | -0.1478 | Yes | ||
| 58 | CAP1 | 2650278 | 18491 | -5.943 | -0.0784 | Yes | ||
| 59 | ANXA5 | 1050373 | 18556 | -7.210 | 0.0032 | Yes |

