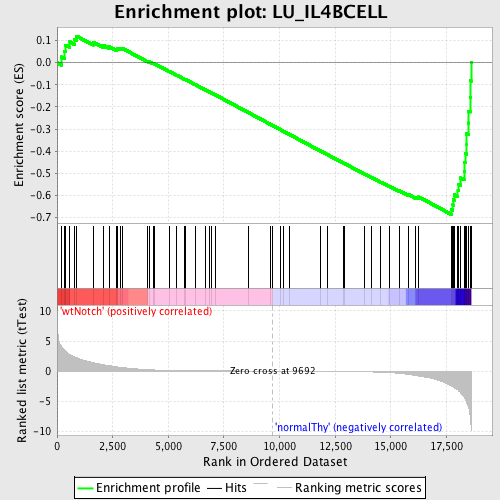
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | LU_IL4BCELL |
| Enrichment Score (ES) | -0.6861718 |
| Normalized Enrichment Score (NES) | -1.6378586 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.18844385 |
| FWER p-Value | 0.733 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | QSCN6 | 3120441 | 204 | 4.025 | 0.0258 | No | ||
| 2 | MRPS15 | 3840280 5360504 | 325 | 3.514 | 0.0515 | No | ||
| 3 | GSR | 3520079 | 373 | 3.332 | 0.0794 | No | ||
| 4 | NFIL3 | 4070377 | 569 | 2.751 | 0.0940 | No | ||
| 5 | VCL | 4120487 | 791 | 2.368 | 0.1038 | No | ||
| 6 | SAMSN1 | 2370563 | 871 | 2.230 | 0.1199 | No | ||
| 7 | EIF4G1 | 4070446 | 1619 | 1.417 | 0.0926 | No | ||
| 8 | SPAG1 | 5570022 | 2085 | 1.044 | 0.0771 | No | ||
| 9 | EVI2A | 6840110 | 2341 | 0.917 | 0.0717 | No | ||
| 10 | RPL5 | 840095 | 2661 | 0.732 | 0.0612 | No | ||
| 11 | RPS2 | 1340647 | 2710 | 0.706 | 0.0651 | No | ||
| 12 | IL2RA | 6620450 | 2842 | 0.642 | 0.0639 | No | ||
| 13 | STAT4 | 1660373 3130019 | 2935 | 0.599 | 0.0644 | No | ||
| 14 | WT1 | 3990463 | 4068 | 0.241 | 0.0056 | No | ||
| 15 | BHLHB2 | 7040603 | 4140 | 0.228 | 0.0039 | No | ||
| 16 | CNTN6 | 2630711 | 4326 | 0.202 | -0.0042 | No | ||
| 17 | CD69 | 380167 4730088 | 4356 | 0.198 | -0.0040 | No | ||
| 18 | TLR7 | 3140300 | 5040 | 0.129 | -0.0396 | No | ||
| 19 | RGS6 | 6040601 | 5364 | 0.109 | -0.0560 | No | ||
| 20 | HOXC4 | 1940193 | 5727 | 0.091 | -0.0747 | No | ||
| 21 | CLTC | 6590458 | 5748 | 0.089 | -0.0750 | No | ||
| 22 | LY75 | 1990358 4590136 | 6197 | 0.071 | -0.0984 | No | ||
| 23 | CTGF | 4540577 | 6689 | 0.055 | -0.1244 | No | ||
| 24 | FGL2 | 3170731 | 6866 | 0.051 | -0.1334 | No | ||
| 25 | TLR3 | 6760451 | 6944 | 0.049 | -0.1371 | No | ||
| 26 | MAL | 4280487 4590239 | 7104 | 0.045 | -0.1453 | No | ||
| 27 | KMO | 7000132 | 8595 | 0.017 | -0.2254 | No | ||
| 28 | NCF2 | 540129 2370441 2650133 | 9602 | 0.001 | -0.2796 | No | ||
| 29 | HLX1 | 4250102 | 9683 | 0.000 | -0.2839 | No | ||
| 30 | RAGE | 730102 2120706 7050315 | 10055 | -0.005 | -0.3039 | No | ||
| 31 | IL6 | 380133 | 10184 | -0.007 | -0.3107 | No | ||
| 32 | CFLAR | 2320605 6180440 | 10426 | -0.011 | -0.3236 | No | ||
| 33 | BCKDK | 1940601 3990600 | 11822 | -0.035 | -0.3985 | No | ||
| 34 | TNFRSF11A | 4230703 | 12140 | -0.042 | -0.4152 | No | ||
| 35 | PMAIP1 | 1400717 5360047 5900039 | 12878 | -0.062 | -0.4543 | No | ||
| 36 | ARPC5 | 1740411 3450300 | 12908 | -0.063 | -0.4553 | No | ||
| 37 | MAP3K5 | 6020041 6380162 | 13822 | -0.108 | -0.5035 | No | ||
| 38 | AKAP2 | 5550347 | 14122 | -0.131 | -0.5184 | No | ||
| 39 | PDE7A | 1690746 2190041 | 14549 | -0.173 | -0.5398 | No | ||
| 40 | NEK6 | 3360687 | 14928 | -0.241 | -0.5580 | No | ||
| 41 | SPINT2 | 2630039 6110519 | 15387 | -0.365 | -0.5793 | No | ||
| 42 | CD86 | 3390471 6200280 | 15804 | -0.543 | -0.5968 | No | ||
| 43 | CTSD | 1070195 | 16128 | -0.728 | -0.6075 | No | ||
| 44 | SIAH2 | 7210672 | 16255 | -0.807 | -0.6070 | No | ||
| 45 | NCOA3 | 4540195 | 17726 | -2.466 | -0.6636 | Yes | ||
| 46 | GNG2 | 2230390 | 17790 | -2.614 | -0.6431 | Yes | ||
| 47 | STK4 | 2640152 | 17801 | -2.623 | -0.6197 | Yes | ||
| 48 | SLA | 2000070 | 17846 | -2.731 | -0.5971 | Yes | ||
| 49 | BATF | 6760390 | 18018 | -3.119 | -0.5778 | Yes | ||
| 50 | PPP2CA | 3990113 | 18056 | -3.288 | -0.5497 | Yes | ||
| 51 | SLC15A2 | 1780538 | 18124 | -3.531 | -0.5210 | Yes | ||
| 52 | SELL | 1190500 | 18294 | -4.327 | -0.4906 | Yes | ||
| 53 | KLF13 | 4670563 | 18309 | -4.420 | -0.4509 | Yes | ||
| 54 | XBP1 | 3840594 | 18340 | -4.607 | -0.4104 | Yes | ||
| 55 | BCL6 | 940100 | 18397 | -5.006 | -0.3677 | Yes | ||
| 56 | PPP6C | 2450670 | 18401 | -5.045 | -0.3217 | Yes | ||
| 57 | CISH | 840315 | 18477 | -5.810 | -0.2726 | Yes | ||
| 58 | IL4R | 2370520 | 18488 | -5.911 | -0.2191 | Yes | ||
| 59 | TNFAIP3 | 2900142 | 18562 | -7.402 | -0.1554 | Yes | ||
| 60 | CD79B | 1450066 3390358 | 18583 | -8.170 | -0.0818 | Yes | ||
| 61 | EGR1 | 4610347 | 18609 | -9.130 | 0.0004 | Yes |

