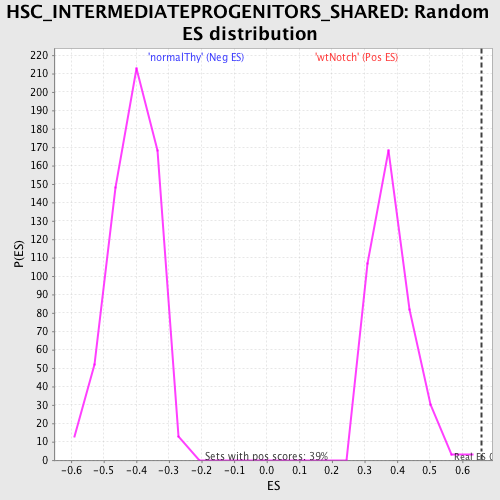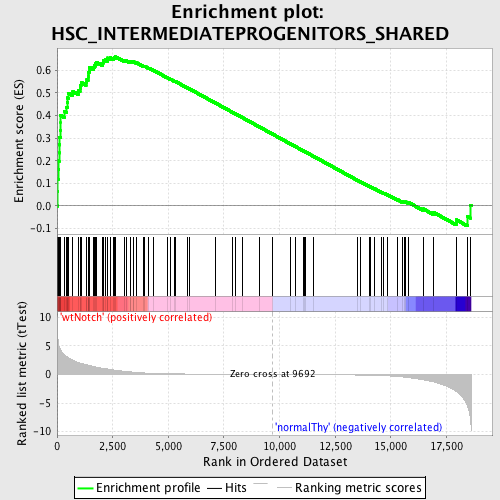
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | wtNotch |
| GeneSet | HSC_INTERMEDIATEPROGENITORS_SHARED |
| Enrichment Score (ES) | 0.65959144 |
| Normalized Enrichment Score (NES) | 1.726777 |
| Nominal p-value | 0.0025445293 |
| FDR q-value | 0.027246956 |
| FWER p-Value | 0.199 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SDC4 | 6370411 | 11 | 8.687 | 0.0645 | Yes | ||
| 2 | C12ORF10 | 4120324 5270113 | 20 | 7.378 | 0.1193 | Yes | ||
| 3 | ALDOA | 6290672 | 47 | 5.798 | 0.1613 | Yes | ||
| 4 | TBRG4 | 3610239 6370594 | 69 | 5.246 | 0.1995 | Yes | ||
| 5 | NPC2 | 2350102 | 90 | 4.819 | 0.2345 | Yes | ||
| 6 | APEX1 | 3190519 | 96 | 4.789 | 0.2701 | Yes | ||
| 7 | GTF3A | 2340451 | 106 | 4.687 | 0.3047 | Yes | ||
| 8 | SDF2L1 | 4230446 | 143 | 4.423 | 0.3359 | Yes | ||
| 9 | CHCHD4 | 2680075 | 151 | 4.320 | 0.3678 | Yes | ||
| 10 | GALK1 | 840162 | 155 | 4.303 | 0.3999 | Yes | ||
| 11 | MRPL12 | 3120112 | 320 | 3.528 | 0.4175 | Yes | ||
| 12 | APIP | 2480440 2810471 | 424 | 3.133 | 0.4354 | Yes | ||
| 13 | ITGB4BP | 1400541 | 456 | 3.045 | 0.4565 | Yes | ||
| 14 | BID | 4590592 6130008 | 476 | 2.990 | 0.4779 | Yes | ||
| 15 | DDX52 | 2970411 | 509 | 2.917 | 0.4980 | Yes | ||
| 16 | DBI | 1090706 | 708 | 2.524 | 0.5062 | Yes | ||
| 17 | COMMD10 | 3120451 | 948 | 2.103 | 0.5091 | Yes | ||
| 18 | GFM1 | 6220711 | 1031 | 1.992 | 0.5196 | Yes | ||
| 19 | KLHDC4 | 870288 5900092 | 1051 | 1.966 | 0.5333 | Yes | ||
| 20 | EEF1B2 | 4850400 6130161 | 1078 | 1.937 | 0.5464 | Yes | ||
| 21 | MAPRE2 | 1660435 5860403 | 1301 | 1.715 | 0.5472 | Yes | ||
| 22 | POLR3E | 380152 | 1338 | 1.679 | 0.5579 | Yes | ||
| 23 | THOC5 | 4570047 | 1392 | 1.635 | 0.5673 | Yes | ||
| 24 | MRPS35 | 2370603 4070551 | 1397 | 1.629 | 0.5792 | Yes | ||
| 25 | GPT2 | 1850463 | 1402 | 1.627 | 0.5912 | Yes | ||
| 26 | SSR2 | 5390180 | 1441 | 1.591 | 0.6011 | Yes | ||
| 27 | TEX2 | 1740504 2480102 3060494 | 1448 | 1.587 | 0.6126 | Yes | ||
| 28 | PES1 | 2470427 | 1642 | 1.402 | 0.6127 | Yes | ||
| 29 | MRPS25 | 610441 870128 1410368 | 1672 | 1.374 | 0.6214 | Yes | ||
| 30 | DDX20 | 3290348 | 1720 | 1.332 | 0.6289 | Yes | ||
| 31 | GMPPA | 520450 3120497 | 1764 | 1.297 | 0.6363 | Yes | ||
| 32 | SURF6 | 3060451 | 2041 | 1.071 | 0.6294 | Yes | ||
| 33 | HES6 | 540411 6550504 | 2065 | 1.057 | 0.6361 | Yes | ||
| 34 | WDR34 | 6100215 | 2095 | 1.040 | 0.6423 | Yes | ||
| 35 | PTCD2 | 1230088 1400605 3170576 6200609 | 2160 | 1.002 | 0.6464 | Yes | ||
| 36 | CTSZ | 1500687 1690364 | 2266 | 0.970 | 0.6480 | Yes | ||
| 37 | LMAN2 | 430154 | 2277 | 0.961 | 0.6546 | Yes | ||
| 38 | TXNDC4 | 450537 | 2386 | 0.890 | 0.6555 | Yes | ||
| 39 | PLAC8 | 2900019 | 2533 | 0.801 | 0.6536 | Yes | ||
| 40 | DHX37 | 1400309 | 2571 | 0.776 | 0.6574 | Yes | ||
| 41 | RSU1 | 4850372 | 2634 | 0.742 | 0.6596 | Yes | ||
| 42 | CPA3 | 610242 6380102 | 3040 | 0.558 | 0.6419 | No | ||
| 43 | CFDP1 | 7000112 | 3113 | 0.532 | 0.6420 | No | ||
| 44 | PCCA | 3390400 | 3287 | 0.464 | 0.6361 | No | ||
| 45 | MAPKAPK3 | 630520 | 3289 | 0.462 | 0.6395 | No | ||
| 46 | TCN2 | 2810139 | 3418 | 0.423 | 0.6358 | No | ||
| 47 | POLR3C | 6940546 | 3432 | 0.416 | 0.6382 | No | ||
| 48 | GCNT3 | 6290239 | 3563 | 0.371 | 0.6340 | No | ||
| 49 | TNS3 | 6380242 | 3903 | 0.276 | 0.6178 | No | ||
| 50 | CDADC1 | 3710201 4120064 | 3930 | 0.269 | 0.6184 | No | ||
| 51 | AP3S2 | 2630411 5080053 | 4116 | 0.233 | 0.6101 | No | ||
| 52 | SIGLEC5 | 5690398 | 4318 | 0.202 | 0.6008 | No | ||
| 53 | CEBPZ | 2970053 2680068 | 4941 | 0.137 | 0.5682 | No | ||
| 54 | MDH1 | 6660358 6760731 | 5112 | 0.124 | 0.5600 | No | ||
| 55 | C5ORF14 | 3780050 | 5277 | 0.114 | 0.5520 | No | ||
| 56 | MAOA | 1410039 4610324 | 5328 | 0.111 | 0.5501 | No | ||
| 57 | PFKP | 70138 6760040 1170278 | 5877 | 0.084 | 0.5212 | No | ||
| 58 | ATP6AP2 | 4010059 5080315 7100347 6660484 | 5971 | 0.080 | 0.5168 | No | ||
| 59 | GPR180 | 5390711 | 7137 | 0.044 | 0.4542 | No | ||
| 60 | DTX4 | 4120433 | 7876 | 0.030 | 0.4146 | No | ||
| 61 | PLAA | 60706 6370369 | 8017 | 0.027 | 0.4073 | No | ||
| 62 | NEBL | 1850369 2480286 | 8347 | 0.021 | 0.3897 | No | ||
| 63 | CITED2 | 5670114 5130088 | 9113 | 0.009 | 0.3484 | No | ||
| 64 | RPN1 | 1240113 | 9669 | 0.000 | 0.3185 | No | ||
| 65 | ELOVL3 | 6860050 | 10482 | -0.011 | 0.2748 | No | ||
| 66 | AS3MT | 1570064 2350113 | 10734 | -0.015 | 0.2613 | No | ||
| 67 | NAT12 | 1780092 4730370 | 11075 | -0.021 | 0.2431 | No | ||
| 68 | GYS1 | 540154 | 11102 | -0.021 | 0.2419 | No | ||
| 69 | BET1 | 4010725 | 11155 | -0.022 | 0.2393 | No | ||
| 70 | AHCY | 4230605 | 11518 | -0.029 | 0.2199 | No | ||
| 71 | STATIP1 | 4480022 6940097 | 13509 | -0.088 | 0.1132 | No | ||
| 72 | CUL5 | 450142 | 13653 | -0.096 | 0.1062 | No | ||
| 73 | NIPSNAP3B | 630672 3710091 4540288 4780576 | 14022 | -0.122 | 0.0872 | No | ||
| 74 | D4ST1 | 1780671 | 14102 | -0.128 | 0.0839 | No | ||
| 75 | HDGF | 5130114 5420066 6590152 | 14275 | -0.146 | 0.0758 | No | ||
| 76 | ATP6V1F | 5420082 | 14588 | -0.178 | 0.0603 | No | ||
| 77 | SCARB1 | 510441 6770047 | 14676 | -0.193 | 0.0570 | No | ||
| 78 | SNUPN | 5050131 | 14858 | -0.226 | 0.0489 | No | ||
| 79 | RASSF4 | 510372 1660673 6290040 | 15317 | -0.343 | 0.0268 | No | ||
| 80 | PAPSS2 | 870113 | 15511 | -0.416 | 0.0195 | No | ||
| 81 | SFXN4 | 60750 4560059 | 15621 | -0.459 | 0.0170 | No | ||
| 82 | NUP37 | 2370097 6370435 6380008 | 15676 | -0.487 | 0.0178 | No | ||
| 83 | ERCC8 | 1240300 6450372 6590180 | 15791 | -0.537 | 0.0156 | No | ||
| 84 | DYNLL1 | 5570113 | 16457 | -0.935 | -0.0133 | No | ||
| 85 | BLOC1S2 | 1240215 | 16928 | -1.272 | -0.0291 | No | ||
| 86 | PYCARD | 2100750 | 17945 | -2.968 | -0.0617 | No | ||
| 87 | SCIN | 5420180 | 18445 | -5.529 | -0.0472 | No | ||
| 88 | CASP7 | 4210156 | 18566 | -7.530 | 0.0027 | No |