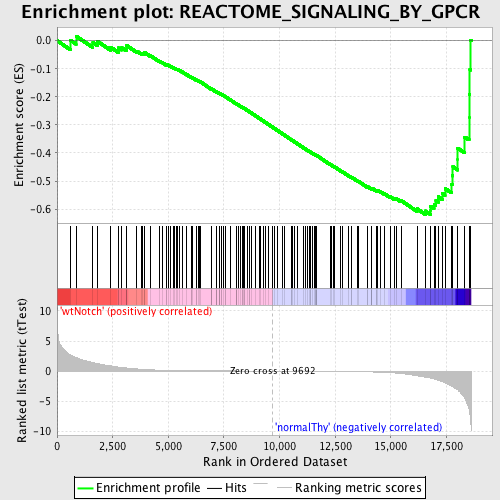
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | REACTOME_SIGNALING_BY_GPCR |
| Enrichment Score (ES) | -0.6191301 |
| Normalized Enrichment Score (NES) | -1.6389548 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.06859014 |
| FWER p-Value | 0.495 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CNR2 | 2760398 | 600 | 2.694 | 0.0014 | No | ||
| 2 | F2 | 5720280 | 852 | 2.255 | 0.0162 | No | ||
| 3 | P2RY6 | 5290400 | 1610 | 1.422 | -0.0069 | No | ||
| 4 | GAL | 3930279 | 1828 | 1.246 | -0.0029 | No | ||
| 5 | P2RY10 | 6370039 6840204 | 2410 | 0.875 | -0.0234 | No | ||
| 6 | P2RY2 | 2640180 2810128 | 2759 | 0.674 | -0.0337 | No | ||
| 7 | GPR17 | 5890671 | 2770 | 0.670 | -0.0258 | No | ||
| 8 | ROCK1 | 130044 | 2909 | 0.612 | -0.0256 | No | ||
| 9 | CCL25 | 450541 540435 | 3111 | 0.533 | -0.0298 | No | ||
| 10 | ADCY6 | 450364 6290670 6940286 | 3129 | 0.525 | -0.0241 | No | ||
| 11 | HRH2 | 3840440 | 3131 | 0.524 | -0.0176 | No | ||
| 12 | SCTR | 2940070 | 3575 | 0.367 | -0.0369 | No | ||
| 13 | LTB4R | 1770056 | 3804 | 0.298 | -0.0455 | No | ||
| 14 | PDE4D | 2470528 6660014 | 3848 | 0.288 | -0.0442 | No | ||
| 15 | NTSR1 | 6510136 | 3923 | 0.271 | -0.0448 | No | ||
| 16 | PDYN | 5720408 | 3935 | 0.268 | -0.0420 | No | ||
| 17 | PNOC | 1990114 | 4179 | 0.222 | -0.0524 | No | ||
| 18 | OPN1SW | 6420377 | 4593 | 0.169 | -0.0726 | No | ||
| 19 | AVP | 2100113 | 4724 | 0.156 | -0.0777 | No | ||
| 20 | GRK5 | 1940348 4670053 | 4902 | 0.140 | -0.0855 | No | ||
| 21 | AVPR1A | 2120300 | 4928 | 0.138 | -0.0851 | No | ||
| 22 | ADCYAP1 | 520086 | 4988 | 0.132 | -0.0867 | No | ||
| 23 | GCG | 6220735 | 5111 | 0.125 | -0.0917 | No | ||
| 24 | ADRA1D | 380035 540025 | 5238 | 0.116 | -0.0971 | No | ||
| 25 | PTGER2 | 3170039 | 5288 | 0.114 | -0.0983 | No | ||
| 26 | PLCB2 | 360132 | 5366 | 0.109 | -0.1011 | No | ||
| 27 | IAPP | 4230040 | 5409 | 0.107 | -0.1020 | No | ||
| 28 | GNA14 | 3390017 | 5504 | 0.101 | -0.1058 | No | ||
| 29 | SSTR1 | 2630471 | 5628 | 0.096 | -0.1113 | No | ||
| 30 | KISS1 | 1580398 | 5834 | 0.085 | -0.1213 | No | ||
| 31 | GALR1 | 6020452 | 6020 | 0.078 | -0.1303 | No | ||
| 32 | MC2R | 2970504 | 6038 | 0.077 | -0.1303 | No | ||
| 33 | PTGER3 | 2900739 6770750 | 6096 | 0.075 | -0.1324 | No | ||
| 34 | CGB | 580273 | 6250 | 0.069 | -0.1398 | No | ||
| 35 | NTS | 380300 2120592 | 6276 | 0.068 | -0.1403 | No | ||
| 36 | IL8RB | 450592 1170537 | 6360 | 0.066 | -0.1440 | No | ||
| 37 | CASR | 610504 | 6393 | 0.064 | -0.1449 | No | ||
| 38 | TSHR | 1190687 5270364 | 6424 | 0.063 | -0.1457 | No | ||
| 39 | CCK | 6370368 | 6922 | 0.049 | -0.1720 | No | ||
| 40 | CCR3 | 50427 | 6932 | 0.049 | -0.1719 | No | ||
| 41 | CXCL11 | 1090551 | 6946 | 0.049 | -0.1720 | No | ||
| 42 | HCRTR2 | 2350463 | 7144 | 0.044 | -0.1821 | No | ||
| 43 | PIK3R1 | 4730671 | 7165 | 0.043 | -0.1826 | No | ||
| 44 | TACR3 | 4780017 | 7168 | 0.043 | -0.1822 | No | ||
| 45 | MTNR1A | 380204 | 7306 | 0.041 | -0.1891 | No | ||
| 46 | TACR2 | 1740358 | 7309 | 0.041 | -0.1887 | No | ||
| 47 | HTR1F | 670148 | 7319 | 0.040 | -0.1887 | No | ||
| 48 | LHCGR | 4560465 5860551 | 7378 | 0.039 | -0.1913 | No | ||
| 49 | DRD1 | 430025 | 7409 | 0.039 | -0.1925 | No | ||
| 50 | VIP | 2850647 | 7468 | 0.037 | -0.1951 | No | ||
| 51 | PPY | 2340373 | 7556 | 0.036 | -0.1994 | No | ||
| 52 | GLP2R | 4150577 5290273 7050095 | 7573 | 0.035 | -0.1998 | No | ||
| 53 | AKT1 | 5290746 | 7808 | 0.031 | -0.2121 | No | ||
| 54 | SST | 6590142 | 8046 | 0.027 | -0.2246 | No | ||
| 55 | HCRT | 2970463 | 8060 | 0.026 | -0.2249 | No | ||
| 56 | ADM2 | 4060102 | 8072 | 0.026 | -0.2252 | No | ||
| 57 | CCKBR | 2760128 | 8161 | 0.024 | -0.2297 | No | ||
| 58 | C5AR1 | 4540402 | 8229 | 0.023 | -0.2330 | No | ||
| 59 | GLP1R | 6420528 | 8317 | 0.022 | -0.2374 | No | ||
| 60 | EDNRA | 6900133 | 8319 | 0.022 | -0.2372 | No | ||
| 61 | GNAT1 | 5270048 6100170 | 8381 | 0.021 | -0.2403 | No | ||
| 62 | GRPR | 6020170 | 8427 | 0.020 | -0.2424 | No | ||
| 63 | ADRA2A | 5340520 | 8443 | 0.019 | -0.2430 | No | ||
| 64 | PPYR1 | 1410072 | 8548 | 0.018 | -0.2484 | No | ||
| 65 | VIPR2 | 6520050 | 8628 | 0.016 | -0.2525 | No | ||
| 66 | GNAZ | 6130296 | 8655 | 0.016 | -0.2537 | No | ||
| 67 | GPRC6A | 4560576 | 8758 | 0.014 | -0.2590 | No | ||
| 68 | GHRHR | 3130500 4010154 | 8910 | 0.012 | -0.2671 | No | ||
| 69 | PTGER1 | 6100132 | 9084 | 0.009 | -0.2763 | No | ||
| 70 | PTGFR | 3850373 | 9145 | 0.008 | -0.2795 | No | ||
| 71 | OXT | 3850692 | 9269 | 0.006 | -0.2860 | No | ||
| 72 | GHRH | 4570575 | 9361 | 0.005 | -0.2909 | No | ||
| 73 | BAI3 | 940692 | 9500 | 0.003 | -0.2983 | No | ||
| 74 | ADORA1 | 6370520 | 9664 | 0.000 | -0.3071 | No | ||
| 75 | GIP | 3170364 | 9670 | 0.000 | -0.3074 | No | ||
| 76 | EDN1 | 1770047 | 9776 | -0.001 | -0.3131 | No | ||
| 77 | PIK3CA | 6220129 | 9883 | -0.003 | -0.3188 | No | ||
| 78 | ADCYAP1R1 | 3390020 4280184 5360180 | 10115 | -0.006 | -0.3312 | No | ||
| 79 | HTR6 | 4850022 | 10205 | -0.007 | -0.3359 | No | ||
| 80 | P2RY4 | 630053 | 10528 | -0.012 | -0.3532 | No | ||
| 81 | CRH | 3710301 | 10541 | -0.012 | -0.3537 | No | ||
| 82 | HRH1 | 840100 | 10576 | -0.013 | -0.3554 | No | ||
| 83 | CALCA | 5860167 | 10676 | -0.015 | -0.3606 | No | ||
| 84 | CCR10 | 4050097 | 10819 | -0.017 | -0.3680 | No | ||
| 85 | GNGT1 | 5220156 | 11089 | -0.021 | -0.3823 | No | ||
| 86 | DARC | 3940050 | 11159 | -0.022 | -0.3858 | No | ||
| 87 | TACR1 | 70358 3840411 | 11256 | -0.024 | -0.3907 | No | ||
| 88 | IL8RA | 1580100 | 11347 | -0.026 | -0.3952 | No | ||
| 89 | CHRM5 | 5390333 | 11392 | -0.027 | -0.3973 | No | ||
| 90 | CHRM2 | 870750 | 11399 | -0.027 | -0.3973 | No | ||
| 91 | KNG1 | 6400576 6770347 | 11477 | -0.028 | -0.4011 | No | ||
| 92 | DRD3 | 4780402 | 11577 | -0.030 | -0.4061 | No | ||
| 93 | TAC1 | 7000195 380706 | 11613 | -0.030 | -0.4076 | No | ||
| 94 | GCGR | 6620497 | 11625 | -0.031 | -0.4078 | No | ||
| 95 | LHB | 1450368 | 11638 | -0.031 | -0.4080 | No | ||
| 96 | HCRTR1 | 1580273 | 11670 | -0.031 | -0.4093 | No | ||
| 97 | AGTR1 | 4780524 2680592 | 11674 | -0.031 | -0.4091 | No | ||
| 98 | CALCR | 1690494 | 12290 | -0.045 | -0.4418 | No | ||
| 99 | FSHR | 610041 7040121 | 12317 | -0.046 | -0.4426 | No | ||
| 100 | CGA | 630403 | 12441 | -0.048 | -0.4487 | No | ||
| 101 | CXCR6 | 3190440 | 12460 | -0.049 | -0.4490 | No | ||
| 102 | TSHB | 6770500 | 12756 | -0.058 | -0.4643 | No | ||
| 103 | XCR1 | 5080072 | 12839 | -0.061 | -0.4680 | No | ||
| 104 | PTGDR | 3850161 | 13104 | -0.070 | -0.4814 | No | ||
| 105 | FSHB | 7100592 | 13233 | -0.075 | -0.4874 | No | ||
| 106 | AGT | 7000575 | 13253 | -0.076 | -0.4874 | No | ||
| 107 | MC5R | 2640176 | 13519 | -0.089 | -0.5007 | No | ||
| 108 | NPFF | 2030280 | 13567 | -0.091 | -0.5021 | No | ||
| 109 | BDKRB1 | 730592 | 13933 | -0.115 | -0.5204 | No | ||
| 110 | OPRM1 | 5360279 | 13964 | -0.118 | -0.5205 | No | ||
| 111 | P2RY1 | 6040121 | 14139 | -0.133 | -0.5283 | No | ||
| 112 | CRHR2 | 4590672 | 14144 | -0.134 | -0.5268 | No | ||
| 113 | ADORA2A | 1990687 | 14147 | -0.134 | -0.5252 | No | ||
| 114 | CALCRL | 4280035 | 14365 | -0.155 | -0.5350 | No | ||
| 115 | CCR6 | 5720368 6020176 | 14371 | -0.156 | -0.5333 | No | ||
| 116 | GRM6 | 2030398 | 14384 | -0.157 | -0.5320 | No | ||
| 117 | CX3CL1 | 3990707 | 14550 | -0.173 | -0.5387 | No | ||
| 118 | GNAQ | 430670 4210131 5900736 | 14693 | -0.195 | -0.5440 | No | ||
| 119 | AVPR2 | 360064 | 14963 | -0.250 | -0.5554 | No | ||
| 120 | GNA12 | 1230301 | 15157 | -0.292 | -0.5622 | No | ||
| 121 | HTR2A | 360037 6330255 | 15252 | -0.327 | -0.5631 | No | ||
| 122 | SCT | 2230348 | 15459 | -0.388 | -0.5694 | No | ||
| 123 | P2RY14 | 6100497 | 16180 | -0.758 | -0.5988 | No | ||
| 124 | GNAS | 630441 1850373 4050152 | 16556 | -0.998 | -0.6066 | Yes | ||
| 125 | GNA13 | 4590102 | 16772 | -1.147 | -0.6038 | Yes | ||
| 126 | PIK3CG | 5890110 | 16778 | -1.152 | -0.5896 | Yes | ||
| 127 | NMB | 3360735 | 16942 | -1.289 | -0.5822 | Yes | ||
| 128 | ADRBK1 | 1340333 | 17027 | -1.377 | -0.5694 | Yes | ||
| 129 | PTGIR | 2100563 | 17132 | -1.521 | -0.5559 | Yes | ||
| 130 | RHOA | 580142 5900131 5340450 | 17313 | -1.730 | -0.5439 | Yes | ||
| 131 | RHOG | 6760575 | 17437 | -1.919 | -0.5264 | Yes | ||
| 132 | F2R | 4810180 | 17732 | -2.480 | -0.5111 | Yes | ||
| 133 | RAMP1 | 2320168 | 17760 | -2.546 | -0.4805 | Yes | ||
| 134 | GNG2 | 2230390 | 17790 | -2.614 | -0.4492 | Yes | ||
| 135 | GNB1 | 2120397 | 18005 | -3.100 | -0.4218 | Yes | ||
| 136 | ADRB2 | 3290373 | 18009 | -3.105 | -0.3829 | Yes | ||
| 137 | PDPK1 | 6650168 | 18323 | -4.496 | -0.3433 | Yes | ||
| 138 | TBXA2R | 2370292 3940066 | 18518 | -6.411 | -0.2731 | Yes | ||
| 139 | XCL1 | 3800504 | 18523 | -6.528 | -0.1912 | Yes | ||
| 140 | CCR7 | 2060687 | 18554 | -7.168 | -0.1026 | Yes | ||
| 141 | CCL5 | 3710397 | 18593 | -8.422 | 0.0012 | Yes |

