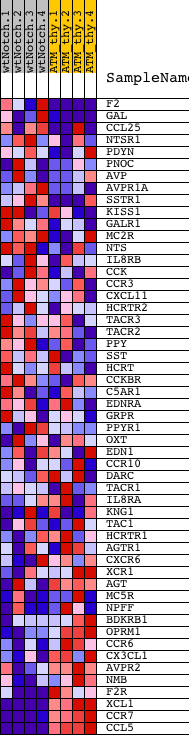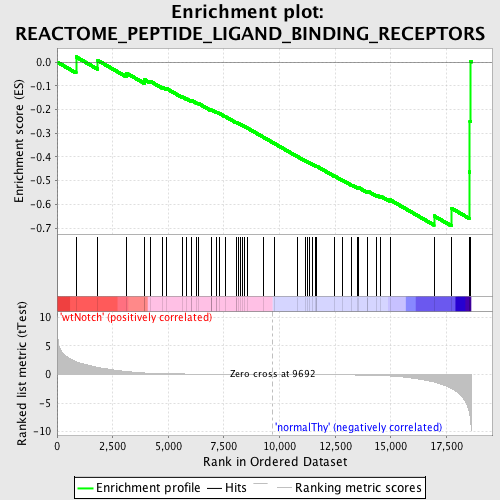
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS |
| Enrichment Score (ES) | -0.69097877 |
| Normalized Enrichment Score (NES) | -1.5747913 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.11780495 |
| FWER p-Value | 0.892 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | F2 | 5720280 | 852 | 2.255 | 0.0218 | No | ||
| 2 | GAL | 3930279 | 1828 | 1.246 | 0.0067 | No | ||
| 3 | CCL25 | 450541 540435 | 3111 | 0.533 | -0.0464 | No | ||
| 4 | NTSR1 | 6510136 | 3923 | 0.271 | -0.0819 | No | ||
| 5 | PDYN | 5720408 | 3935 | 0.268 | -0.0745 | No | ||
| 6 | PNOC | 1990114 | 4179 | 0.222 | -0.0809 | No | ||
| 7 | AVP | 2100113 | 4724 | 0.156 | -0.1055 | No | ||
| 8 | AVPR1A | 2120300 | 4928 | 0.138 | -0.1123 | No | ||
| 9 | SSTR1 | 2630471 | 5628 | 0.096 | -0.1471 | No | ||
| 10 | KISS1 | 1580398 | 5834 | 0.085 | -0.1556 | No | ||
| 11 | GALR1 | 6020452 | 6020 | 0.078 | -0.1632 | No | ||
| 12 | MC2R | 2970504 | 6038 | 0.077 | -0.1618 | No | ||
| 13 | NTS | 380300 2120592 | 6276 | 0.068 | -0.1725 | No | ||
| 14 | IL8RB | 450592 1170537 | 6360 | 0.066 | -0.1750 | No | ||
| 15 | CCK | 6370368 | 6922 | 0.049 | -0.2038 | No | ||
| 16 | CCR3 | 50427 | 6932 | 0.049 | -0.2028 | No | ||
| 17 | CXCL11 | 1090551 | 6946 | 0.049 | -0.2020 | No | ||
| 18 | HCRTR2 | 2350463 | 7144 | 0.044 | -0.2113 | No | ||
| 19 | TACR3 | 4780017 | 7168 | 0.043 | -0.2112 | No | ||
| 20 | TACR2 | 1740358 | 7309 | 0.041 | -0.2176 | No | ||
| 21 | PPY | 2340373 | 7556 | 0.036 | -0.2297 | No | ||
| 22 | SST | 6590142 | 8046 | 0.027 | -0.2553 | No | ||
| 23 | HCRT | 2970463 | 8060 | 0.026 | -0.2552 | No | ||
| 24 | CCKBR | 2760128 | 8161 | 0.024 | -0.2599 | No | ||
| 25 | C5AR1 | 4540402 | 8229 | 0.023 | -0.2628 | No | ||
| 26 | EDNRA | 6900133 | 8319 | 0.022 | -0.2669 | No | ||
| 27 | GRPR | 6020170 | 8427 | 0.020 | -0.2721 | No | ||
| 28 | PPYR1 | 1410072 | 8548 | 0.018 | -0.2780 | No | ||
| 29 | OXT | 3850692 | 9269 | 0.006 | -0.3166 | No | ||
| 30 | EDN1 | 1770047 | 9776 | -0.001 | -0.3438 | No | ||
| 31 | CCR10 | 4050097 | 10819 | -0.017 | -0.3995 | No | ||
| 32 | DARC | 3940050 | 11159 | -0.022 | -0.4170 | No | ||
| 33 | TACR1 | 70358 3840411 | 11256 | -0.024 | -0.4215 | No | ||
| 34 | IL8RA | 1580100 | 11347 | -0.026 | -0.4256 | No | ||
| 35 | KNG1 | 6400576 6770347 | 11477 | -0.028 | -0.4317 | No | ||
| 36 | TAC1 | 7000195 380706 | 11613 | -0.030 | -0.4381 | No | ||
| 37 | HCRTR1 | 1580273 | 11670 | -0.031 | -0.4401 | No | ||
| 38 | AGTR1 | 4780524 2680592 | 11674 | -0.031 | -0.4393 | No | ||
| 39 | CXCR6 | 3190440 | 12460 | -0.049 | -0.4802 | No | ||
| 40 | XCR1 | 5080072 | 12839 | -0.061 | -0.4987 | No | ||
| 41 | AGT | 7000575 | 13253 | -0.076 | -0.5187 | No | ||
| 42 | MC5R | 2640176 | 13519 | -0.089 | -0.5303 | No | ||
| 43 | NPFF | 2030280 | 13567 | -0.091 | -0.5301 | No | ||
| 44 | BDKRB1 | 730592 | 13933 | -0.115 | -0.5463 | No | ||
| 45 | OPRM1 | 5360279 | 13964 | -0.118 | -0.5444 | No | ||
| 46 | CCR6 | 5720368 6020176 | 14371 | -0.156 | -0.5616 | No | ||
| 47 | CX3CL1 | 3990707 | 14550 | -0.173 | -0.5659 | No | ||
| 48 | AVPR2 | 360064 | 14963 | -0.250 | -0.5806 | No | ||
| 49 | NMB | 3360735 | 16942 | -1.289 | -0.6485 | Yes | ||
| 50 | F2R | 4810180 | 17732 | -2.480 | -0.6165 | Yes | ||
| 51 | XCL1 | 3800504 | 18523 | -6.528 | -0.4631 | Yes | ||
| 52 | CCR7 | 2060687 | 18554 | -7.168 | -0.2495 | Yes | ||
| 53 | CCL5 | 3710397 | 18593 | -8.422 | 0.0012 | Yes |