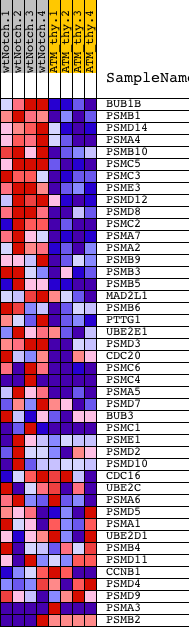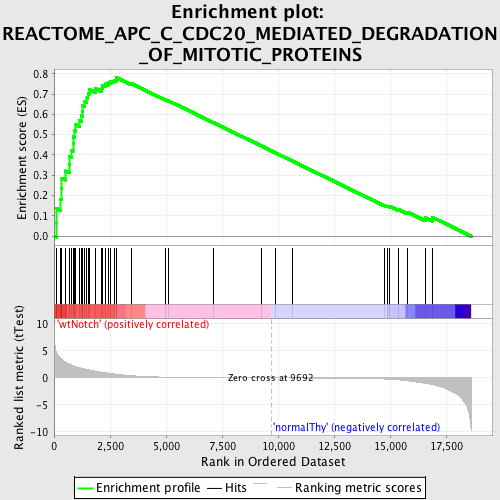
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | wtNotch |
| GeneSet | REACTOME_APC_C_CDC20_MEDIATED_DEGRADATION_OF_MITOTIC_PROTEINS |
| Enrichment Score (ES) | 0.7809731 |
| Normalized Enrichment Score (NES) | 1.8195853 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0036206467 |
| FWER p-Value | 0.0070 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BUB1B | 1450288 | 117 | 4.603 | 0.0649 | Yes | ||
| 2 | PSMB1 | 2940402 | 120 | 4.592 | 0.1359 | Yes | ||
| 3 | PSMD14 | 5690593 | 297 | 3.628 | 0.1825 | Yes | ||
| 4 | PSMA4 | 4560427 | 344 | 3.426 | 0.2331 | Yes | ||
| 5 | PSMB10 | 630504 6510039 | 349 | 3.408 | 0.2856 | Yes | ||
| 6 | PSMC5 | 2760315 6550021 | 519 | 2.882 | 0.3211 | Yes | ||
| 7 | PSMC3 | 2480184 | 677 | 2.568 | 0.3524 | Yes | ||
| 8 | PSME3 | 2810537 | 703 | 2.529 | 0.3902 | Yes | ||
| 9 | PSMD12 | 730044 | 790 | 2.371 | 0.4223 | Yes | ||
| 10 | PSMD8 | 630142 | 846 | 2.266 | 0.4544 | Yes | ||
| 11 | PSMC2 | 7040010 | 854 | 2.254 | 0.4889 | Yes | ||
| 12 | PSMA7 | 2230131 | 925 | 2.151 | 0.5184 | Yes | ||
| 13 | PSMA2 | 6510093 | 973 | 2.074 | 0.5480 | Yes | ||
| 14 | PSMB9 | 6980471 | 1130 | 1.882 | 0.5687 | Yes | ||
| 15 | PSMB3 | 4280594 | 1211 | 1.799 | 0.5922 | Yes | ||
| 16 | PSMB5 | 6290242 | 1272 | 1.738 | 0.6159 | Yes | ||
| 17 | MAD2L1 | 4480725 | 1285 | 1.731 | 0.6420 | Yes | ||
| 18 | PSMB6 | 2690711 | 1374 | 1.653 | 0.6629 | Yes | ||
| 19 | PTTG1 | 520463 | 1467 | 1.567 | 0.6822 | Yes | ||
| 20 | UBE2E1 | 4230064 1090440 | 1512 | 1.517 | 0.7033 | Yes | ||
| 21 | PSMD3 | 1400647 | 1574 | 1.461 | 0.7226 | Yes | ||
| 22 | CDC20 | 3440017 3440044 6220088 | 1858 | 1.226 | 0.7263 | Yes | ||
| 23 | PSMC6 | 2810021 | 2104 | 1.035 | 0.7292 | Yes | ||
| 24 | PSMC4 | 580050 1580025 | 2169 | 1.001 | 0.7412 | Yes | ||
| 25 | PSMA5 | 5390537 | 2289 | 0.953 | 0.7495 | Yes | ||
| 26 | PSMD7 | 2030619 6220594 | 2407 | 0.876 | 0.7568 | Yes | ||
| 27 | BUB3 | 3170546 | 2507 | 0.819 | 0.7641 | Yes | ||
| 28 | PSMC1 | 6350538 | 2674 | 0.725 | 0.7664 | Yes | ||
| 29 | PSME1 | 450193 4480035 | 2780 | 0.668 | 0.7711 | Yes | ||
| 30 | PSMD2 | 4670706 5050364 | 2788 | 0.664 | 0.7810 | Yes | ||
| 31 | PSMD10 | 520494 1170576 3830050 | 3463 | 0.403 | 0.7509 | No | ||
| 32 | CDC16 | 1940706 | 4964 | 0.134 | 0.6722 | No | ||
| 33 | UBE2C | 6130017 | 5106 | 0.125 | 0.6666 | No | ||
| 34 | PSMA6 | 50609 | 7110 | 0.045 | 0.5594 | No | ||
| 35 | PSMD5 | 3940139 4570041 | 9264 | 0.006 | 0.4436 | No | ||
| 36 | PSMA1 | 380059 2760195 | 9875 | -0.002 | 0.4108 | No | ||
| 37 | UBE2D1 | 1850400 | 10642 | -0.014 | 0.3698 | No | ||
| 38 | PSMB4 | 520402 | 14735 | -0.202 | 0.1526 | No | ||
| 39 | PSMD11 | 2340538 6510053 | 14886 | -0.231 | 0.1481 | No | ||
| 40 | CCNB1 | 4590433 4780372 | 14993 | -0.257 | 0.1463 | No | ||
| 41 | PSMD4 | 430068 | 15355 | -0.356 | 0.1324 | No | ||
| 42 | PSMD9 | 50020 5890368 | 15775 | -0.532 | 0.1181 | No | ||
| 43 | PSMA3 | 5900047 7040161 | 16565 | -1.000 | 0.0911 | No | ||
| 44 | PSMB2 | 940035 4210324 | 16898 | -1.247 | 0.0925 | No |