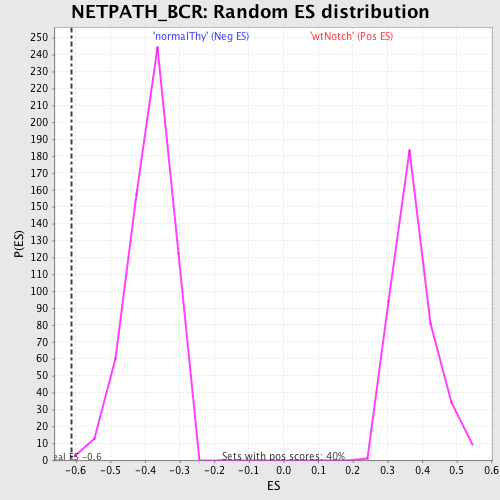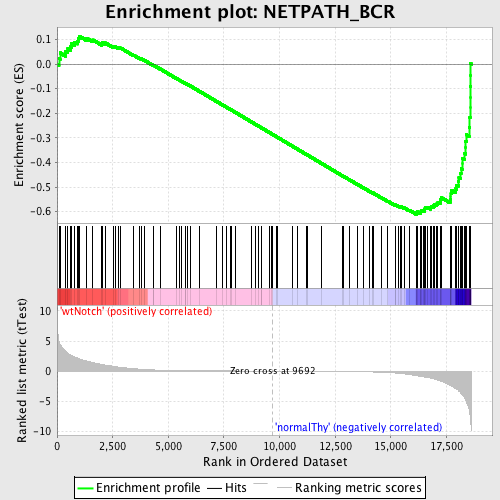
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | NETPATH_BCR |
| Enrichment Score (ES) | -0.61168903 |
| Normalized Enrichment Score (NES) | -1.5880983 |
| Nominal p-value | 0.0016722408 |
| FDR q-value | 0.112230085 |
| FWER p-Value | 0.834 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BAX | 3830008 | 85 | 4.907 | 0.0231 | No | ||
| 2 | RPS6 | 2060168 | 135 | 4.476 | 0.0458 | No | ||
| 3 | CYCS | 2030072 | 384 | 3.284 | 0.0509 | No | ||
| 4 | PIP5K1C | 5890164 6510148 | 484 | 2.978 | 0.0624 | No | ||
| 5 | MAPKAPK2 | 2850500 | 616 | 2.672 | 0.0704 | No | ||
| 6 | PIK3R2 | 1660193 | 656 | 2.603 | 0.0830 | No | ||
| 7 | RPS6KA1 | 4120647 | 789 | 2.372 | 0.0892 | No | ||
| 8 | PTK2 | 1780148 | 928 | 2.143 | 0.0939 | No | ||
| 9 | GRB2 | 6650398 | 957 | 2.091 | 0.1042 | No | ||
| 10 | FCGR2B | 780750 | 993 | 2.050 | 0.1139 | No | ||
| 11 | PTPRC | 130402 5290148 | 1316 | 1.706 | 0.1061 | No | ||
| 12 | PRKCD | 770592 | 1612 | 1.421 | 0.0982 | No | ||
| 13 | ITPR2 | 5360128 | 2007 | 1.100 | 0.0831 | No | ||
| 14 | GSK3A | 3140735 | 2035 | 1.074 | 0.0877 | No | ||
| 15 | PPP3R1 | 1190041 1570487 | 2170 | 1.000 | 0.0861 | No | ||
| 16 | DAPP1 | 2480086 2970300 4480494 | 2532 | 0.801 | 0.0711 | No | ||
| 17 | HCLS1 | 7100309 | 2608 | 0.756 | 0.0713 | No | ||
| 18 | MAP3K7 | 6040068 | 2753 | 0.679 | 0.0674 | No | ||
| 19 | SOS2 | 2060722 | 2853 | 0.638 | 0.0656 | No | ||
| 20 | ZAP70 | 1410494 2260504 | 3430 | 0.417 | 0.0369 | No | ||
| 21 | PPP3CA | 4760332 6760092 | 3702 | 0.323 | 0.0240 | No | ||
| 22 | CCNA2 | 5290075 | 3772 | 0.303 | 0.0220 | No | ||
| 23 | REL | 360707 | 3934 | 0.268 | 0.0148 | No | ||
| 24 | RPS6KB1 | 1450427 5080110 6200563 | 4328 | 0.202 | -0.0053 | No | ||
| 25 | NFATC1 | 510400 2320348 4050600 6180161 6290136 6620086 | 4337 | 0.200 | -0.0046 | No | ||
| 26 | SOS1 | 7050338 | 4633 | 0.165 | -0.0196 | No | ||
| 27 | NFATC2 | 70450 540097 | 5380 | 0.108 | -0.0593 | No | ||
| 28 | RAPGEF1 | 60040 4590670 | 5505 | 0.101 | -0.0654 | No | ||
| 29 | CREB1 | 1500717 2230358 3610600 6550601 | 5595 | 0.097 | -0.0697 | No | ||
| 30 | CD72 | 2340369 | 5779 | 0.087 | -0.0791 | No | ||
| 31 | LYN | 6040600 | 5783 | 0.087 | -0.0787 | No | ||
| 32 | RASA1 | 1240315 | 5862 | 0.084 | -0.0825 | No | ||
| 33 | DOK3 | 130347 3870167 | 6004 | 0.078 | -0.0897 | No | ||
| 34 | LAT2 | 5340440 | 6402 | 0.064 | -0.1108 | No | ||
| 35 | PIK3R1 | 4730671 | 7165 | 0.043 | -0.1517 | No | ||
| 36 | CD22 | 3120239 | 7416 | 0.038 | -0.1650 | No | ||
| 37 | DOK1 | 1050279 2680112 5220273 | 7603 | 0.035 | -0.1748 | No | ||
| 38 | AKT1 | 5290746 | 7808 | 0.031 | -0.1857 | No | ||
| 39 | HCK | 4230592 | 7826 | 0.031 | -0.1864 | No | ||
| 40 | CDK7 | 2640451 | 8013 | 0.027 | -0.1963 | No | ||
| 41 | BCAR1 | 1340215 | 8745 | 0.015 | -0.2358 | No | ||
| 42 | CBLB | 3520465 4670348 | 8912 | 0.012 | -0.2447 | No | ||
| 43 | ACTR2 | 1170332 | 9061 | 0.009 | -0.2526 | No | ||
| 44 | PTPN11 | 2230100 2470180 6100528 | 9168 | 0.008 | -0.2583 | No | ||
| 45 | PRKCQ | 2260170 3870193 | 9201 | 0.007 | -0.2600 | No | ||
| 46 | GAB2 | 1410280 2340520 4280040 | 9538 | 0.002 | -0.2781 | No | ||
| 47 | NCK1 | 6200575 6510050 | 9651 | 0.001 | -0.2842 | No | ||
| 48 | CASP9 | 60577 1170368 2060576 | 9666 | 0.000 | -0.2849 | No | ||
| 49 | BTK | 3130044 | 9841 | -0.002 | -0.2943 | No | ||
| 50 | MAPK4 | 870142 | 9905 | -0.003 | -0.2977 | No | ||
| 51 | ACTR3 | 1400497 | 10591 | -0.013 | -0.3347 | No | ||
| 52 | CD19 | 6940692 | 10826 | -0.017 | -0.3472 | No | ||
| 53 | PRKCE | 5700053 | 11229 | -0.024 | -0.3688 | No | ||
| 54 | RASGRP3 | 6020504 | 11261 | -0.024 | -0.3704 | No | ||
| 55 | CHUK | 7050736 | 11886 | -0.036 | -0.4039 | No | ||
| 56 | PLCG1 | 6020369 | 12826 | -0.060 | -0.4543 | No | ||
| 57 | BLNK | 70288 6590092 | 12888 | -0.062 | -0.4572 | No | ||
| 58 | PIK3AP1 | 6760008 | 13134 | -0.071 | -0.4701 | No | ||
| 59 | CD79A | 3450563 | 13486 | -0.087 | -0.4886 | No | ||
| 60 | SYK | 6940133 | 13785 | -0.106 | -0.5041 | No | ||
| 61 | CR2 | 2850332 6450397 | 14041 | -0.124 | -0.5172 | No | ||
| 62 | PRKD1 | 1410706 5090438 | 14172 | -0.136 | -0.5234 | No | ||
| 63 | SHC1 | 2900731 3170504 6520537 | 14198 | -0.138 | -0.5240 | No | ||
| 64 | JUN | 840170 | 14564 | -0.175 | -0.5427 | No | ||
| 65 | MAPK1 | 3190193 6200253 | 14867 | -0.227 | -0.5578 | No | ||
| 66 | CDK6 | 4920253 | 15196 | -0.305 | -0.5738 | No | ||
| 67 | VAV2 | 3610725 5890717 | 15209 | -0.312 | -0.5727 | No | ||
| 68 | MAPK3 | 580161 4780035 | 15356 | -0.356 | -0.5785 | No | ||
| 69 | IKBKG | 3450092 3840377 6590592 | 15419 | -0.375 | -0.5798 | No | ||
| 70 | IKBKB | 6840072 | 15462 | -0.391 | -0.5798 | No | ||
| 71 | CCNE1 | 110064 | 15604 | -0.450 | -0.5849 | No | ||
| 72 | ATF2 | 2260348 2480441 | 15822 | -0.552 | -0.5935 | No | ||
| 73 | LCP2 | 2680066 6650707 | 16159 | -0.742 | -0.6075 | Yes | ||
| 74 | CDK4 | 540075 4540600 | 16170 | -0.750 | -0.6038 | Yes | ||
| 75 | MAPK8 | 2640195 | 16187 | -0.761 | -0.6004 | Yes | ||
| 76 | FYN | 2100468 4760520 4850687 | 16323 | -0.850 | -0.6029 | Yes | ||
| 77 | MAP4K1 | 4060692 | 16352 | -0.869 | -0.5995 | Yes | ||
| 78 | PTPN6 | 4230128 | 16387 | -0.891 | -0.5963 | Yes | ||
| 79 | WAS | 5270193 | 16461 | -0.938 | -0.5949 | Yes | ||
| 80 | BLK | 1940128 5390053 | 16522 | -0.981 | -0.5926 | Yes | ||
| 81 | CRK | 1230162 4780128 | 16534 | -0.989 | -0.5876 | Yes | ||
| 82 | CSK | 6350593 | 16555 | -0.997 | -0.5831 | Yes | ||
| 83 | GTF2I | 2470750 3870408 3830369 3990082 4610138 | 16650 | -1.044 | -0.5823 | Yes | ||
| 84 | PTPN18 | 2640537 | 16793 | -1.164 | -0.5834 | Yes | ||
| 85 | PDK2 | 2690017 | 16814 | -1.184 | -0.5778 | Yes | ||
| 86 | BCL2 | 730132 1570736 2470138 3800044 4810037 5690068 5860504 6650164 | 16927 | -1.270 | -0.5767 | Yes | ||
| 87 | CD81 | 5270093 | 16960 | -1.304 | -0.5710 | Yes | ||
| 88 | MAPK14 | 5290731 | 17073 | -1.446 | -0.5689 | Yes | ||
| 89 | CDK2 | 130484 2260301 4010088 5050110 | 17116 | -1.496 | -0.5627 | Yes | ||
| 90 | RELA | 3830075 | 17220 | -1.623 | -0.5591 | Yes | ||
| 91 | HDAC5 | 2370035 6370139 | 17227 | -1.631 | -0.5502 | Yes | ||
| 92 | RB1 | 5900338 | 17268 | -1.683 | -0.5429 | Yes | ||
| 93 | LCK | 3360142 | 17665 | -2.357 | -0.5510 | Yes | ||
| 94 | NFATC3 | 5050520 | 17701 | -2.424 | -0.5392 | Yes | ||
| 95 | GSK3B | 5360348 | 17703 | -2.429 | -0.5255 | Yes | ||
| 96 | SH3BP2 | 610131 | 17723 | -2.463 | -0.5126 | Yes | ||
| 97 | VAV1 | 6020487 | 17923 | -2.904 | -0.5070 | Yes | ||
| 98 | GAB1 | 2970156 | 17961 | -2.990 | -0.4921 | Yes | ||
| 99 | BCL10 | 2360397 | 18036 | -3.190 | -0.4781 | Yes | ||
| 100 | CBL | 6380068 | 18061 | -3.305 | -0.4607 | Yes | ||
| 101 | BCL2L11 | 780044 4200601 | 18141 | -3.609 | -0.4446 | Yes | ||
| 102 | NFKBIA | 1570152 | 18160 | -3.680 | -0.4248 | Yes | ||
| 103 | NEDD9 | 1740373 4230053 | 18207 | -3.858 | -0.4055 | Yes | ||
| 104 | CRKL | 4050427 | 18209 | -3.866 | -0.3837 | Yes | ||
| 105 | PDPK1 | 6650168 | 18323 | -4.496 | -0.3644 | Yes | ||
| 106 | CARD11 | 70338 | 18353 | -4.684 | -0.3395 | Yes | ||
| 107 | CCND3 | 380528 730500 | 18358 | -4.702 | -0.3132 | Yes | ||
| 108 | BCL6 | 940100 | 18397 | -5.006 | -0.2870 | Yes | ||
| 109 | PTK2B | 4730411 | 18521 | -6.456 | -0.2571 | Yes | ||
| 110 | ITK | 2230592 | 18551 | -7.151 | -0.2183 | Yes | ||
| 111 | CASP7 | 4210156 | 18566 | -7.530 | -0.1765 | Yes | ||
| 112 | CD5 | 6590338 | 18568 | -7.559 | -0.1339 | Yes | ||
| 113 | TEC | 1400576 | 18571 | -7.590 | -0.0911 | Yes | ||
| 114 | CD79B | 1450066 3390358 | 18583 | -8.170 | -0.0456 | Yes | ||
| 115 | CCND2 | 5340167 | 18589 | -8.378 | 0.0015 | Yes |