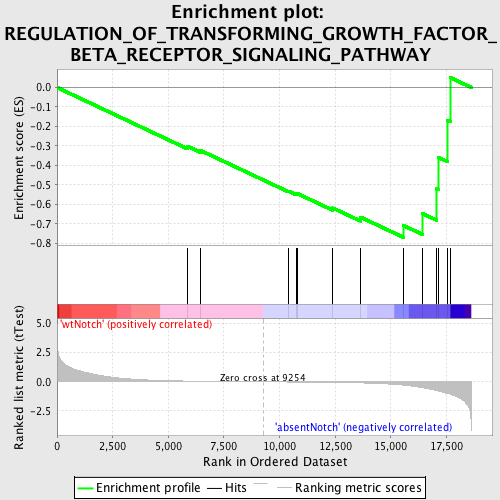
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #wtNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#wtNotch_versus_absentNotch |
| Upregulated in class | absentNotch |
| GeneSet | REGULATION_OF_TRANSFORMING_GROWTH_FACTOR_BETA_RECEPTOR_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.7694868 |
| Normalized Enrichment Score (NES) | -1.5160656 |
| Nominal p-value | 0.030075189 |
| FDR q-value | 0.72248256 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | IL17F | 670577 | 5850 | 0.062 | -0.3012 | No | ||
| 2 | ENG | 4280270 | 6439 | 0.047 | -0.3228 | No | ||
| 3 | SNX6 | 6200086 | 10390 | -0.016 | -0.5317 | No | ||
| 4 | TGFB1 | 1940162 | 10755 | -0.021 | -0.5467 | No | ||
| 5 | CIDEA | 4560020 | 10797 | -0.022 | -0.5442 | No | ||
| 6 | HIPK2 | 6350647 | 12386 | -0.053 | -0.6182 | No | ||
| 7 | SMAD2 | 4200592 | 13635 | -0.094 | -0.6652 | No | ||
| 8 | CDKN2B | 6020040 | 15576 | -0.285 | -0.7086 | Yes | ||
| 9 | CITED2 | 5670114 5130088 | 16426 | -0.502 | -0.6471 | Yes | ||
| 10 | EID2 | 3130176 | 17057 | -0.751 | -0.5206 | Yes | ||
| 11 | SMAD3 | 6450671 | 17125 | -0.778 | -0.3581 | Yes | ||
| 12 | CDKN1C | 6520577 | 17550 | -0.999 | -0.1676 | Yes | ||
| 13 | SMAD4 | 5670519 | 17673 | -1.053 | 0.0507 | Yes |

