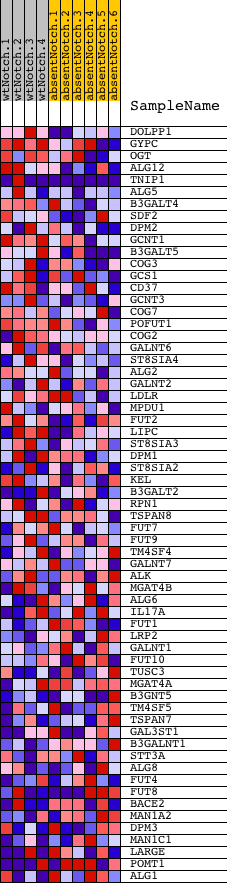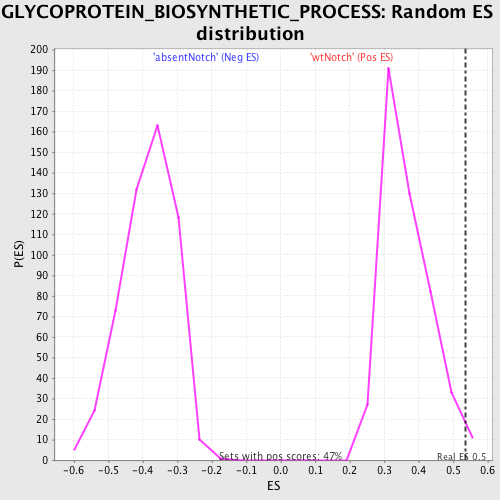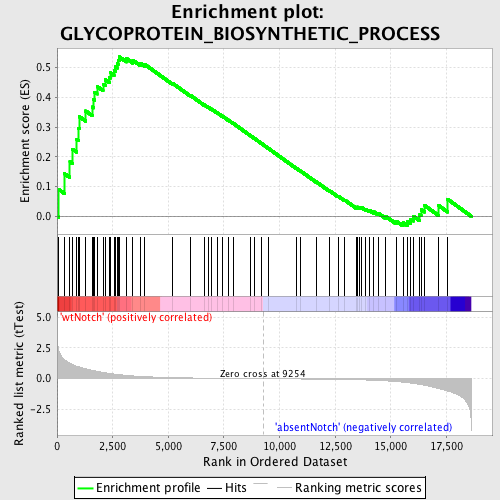
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #wtNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#wtNotch_versus_absentNotch |
| Upregulated in class | wtNotch |
| GeneSet | GLYCOPROTEIN_BIOSYNTHETIC_PROCESS |
| Enrichment Score (ES) | 0.53579605 |
| Normalized Enrichment Score (NES) | 1.4659833 |
| Nominal p-value | 0.021097047 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DOLPP1 | 4780619 6400053 | 83 | 2.200 | 0.0899 | Yes | ||
| 2 | GYPC | 2320102 | 323 | 1.556 | 0.1437 | Yes | ||
| 3 | OGT | 2360131 4610333 | 577 | 1.238 | 0.1832 | Yes | ||
| 4 | ALG12 | 1400692 | 697 | 1.140 | 0.2256 | Yes | ||
| 5 | TNIP1 | 2510279 5900041 6510435 | 883 | 1.007 | 0.2588 | Yes | ||
| 6 | ALG5 | 3060692 4780093 6590097 | 971 | 0.974 | 0.2959 | Yes | ||
| 7 | B3GALT4 | 2340239 | 1005 | 0.946 | 0.3347 | Yes | ||
| 8 | SDF2 | 2690601 | 1284 | 0.800 | 0.3540 | Yes | ||
| 9 | DPM2 | 3610253 | 1577 | 0.681 | 0.3675 | Yes | ||
| 10 | GCNT1 | 940129 1780215 2370253 | 1644 | 0.652 | 0.3919 | Yes | ||
| 11 | B3GALT5 | 4060047 | 1690 | 0.631 | 0.4165 | Yes | ||
| 12 | COG3 | 3840746 | 1795 | 0.599 | 0.4366 | Yes | ||
| 13 | GCS1 | 1740603 | 2065 | 0.512 | 0.4440 | Yes | ||
| 14 | CD37 | 770273 | 2175 | 0.478 | 0.4587 | Yes | ||
| 15 | GCNT3 | 6290239 | 2346 | 0.430 | 0.4680 | Yes | ||
| 16 | COG7 | 360133 | 2411 | 0.410 | 0.4821 | Yes | ||
| 17 | POFUT1 | 1570458 | 2562 | 0.371 | 0.4899 | Yes | ||
| 18 | COG2 | 5900129 | 2623 | 0.361 | 0.5022 | Yes | ||
| 19 | GALNT6 | 3710377 | 2698 | 0.343 | 0.5129 | Yes | ||
| 20 | ST8SIA4 | 2680605 3060215 | 2744 | 0.335 | 0.5248 | Yes | ||
| 21 | ALG2 | 3990091 | 2799 | 0.324 | 0.5358 | Yes | ||
| 22 | GALNT2 | 2260279 4060239 | 3121 | 0.268 | 0.5300 | No | ||
| 23 | LDLR | 5670386 | 3405 | 0.224 | 0.5243 | No | ||
| 24 | MPDU1 | 1850091 | 3760 | 0.183 | 0.5131 | No | ||
| 25 | FUT2 | 3990047 | 3946 | 0.166 | 0.5102 | No | ||
| 26 | LIPC | 2690497 | 5187 | 0.087 | 0.4471 | No | ||
| 27 | ST8SIA3 | 6940427 | 5989 | 0.058 | 0.4064 | No | ||
| 28 | DPM1 | 1240050 4810019 6380398 | 6617 | 0.043 | 0.3745 | No | ||
| 29 | ST8SIA2 | 2680082 | 6798 | 0.039 | 0.3665 | No | ||
| 30 | KEL | 6940440 | 6935 | 0.037 | 0.3607 | No | ||
| 31 | B3GALT2 | 430136 | 7204 | 0.031 | 0.3476 | No | ||
| 32 | RPN1 | 1240113 | 7420 | 0.027 | 0.3372 | No | ||
| 33 | TSPAN8 | 1740538 | 7689 | 0.023 | 0.3237 | No | ||
| 34 | FUT7 | 2900736 | 7919 | 0.019 | 0.3122 | No | ||
| 35 | FUT9 | 3940121 | 8687 | 0.008 | 0.2712 | No | ||
| 36 | TM4SF4 | 540600 | 8871 | 0.005 | 0.2615 | No | ||
| 37 | GALNT7 | 3990280 | 9168 | 0.001 | 0.2456 | No | ||
| 38 | ALK | 730008 | 9515 | -0.004 | 0.2271 | No | ||
| 39 | MGAT4B | 130053 6840100 | 10753 | -0.021 | 0.1614 | No | ||
| 40 | ALG6 | 4060131 | 10944 | -0.025 | 0.1522 | No | ||
| 41 | IL17A | 4280369 | 11639 | -0.037 | 0.1164 | No | ||
| 42 | FUT1 | 1500068 | 12253 | -0.050 | 0.0854 | No | ||
| 43 | LRP2 | 1400204 3290706 | 12636 | -0.059 | 0.0674 | No | ||
| 44 | GALNT1 | 4070487 6130100 6250132 6900048 | 12933 | -0.068 | 0.0543 | No | ||
| 45 | FUT10 | 6510603 | 13461 | -0.087 | 0.0297 | No | ||
| 46 | TUSC3 | 1690411 | 13502 | -0.089 | 0.0313 | No | ||
| 47 | MGAT4A | 5690471 | 13579 | -0.092 | 0.0311 | No | ||
| 48 | B3GNT5 | 4780528 | 13660 | -0.095 | 0.0309 | No | ||
| 49 | TM4SF5 | 3120280 | 13880 | -0.106 | 0.0236 | No | ||
| 50 | TSPAN7 | 870133 | 14030 | -0.115 | 0.0206 | No | ||
| 51 | GAL3ST1 | 5080746 | 14237 | -0.128 | 0.0150 | No | ||
| 52 | B3GALNT1 | 3390746 60131 | 14425 | -0.141 | 0.0109 | No | ||
| 53 | STT3A | 580139 | 14781 | -0.172 | -0.0008 | No | ||
| 54 | ALG8 | 4150497 4780500 6290563 | 15246 | -0.229 | -0.0160 | No | ||
| 55 | FUT4 | 2030324 | 15553 | -0.281 | -0.0205 | No | ||
| 56 | FUT8 | 1340068 2340056 | 15738 | -0.320 | -0.0167 | No | ||
| 57 | BACE2 | 4060170 5690100 6220133 | 15878 | -0.350 | -0.0092 | No | ||
| 58 | MAN1A2 | 5700435 5720114 | 16005 | -0.384 | 0.0005 | No | ||
| 59 | DPM3 | 50039 | 16296 | -0.463 | 0.0048 | No | ||
| 60 | MAN1C1 | 730133 870398 2340044 | 16362 | -0.485 | 0.0220 | No | ||
| 61 | LARGE | 6940068 | 16507 | -0.529 | 0.0370 | No | ||
| 62 | POMT1 | 2680047 3440040 6660114 | 17145 | -0.787 | 0.0364 | No | ||
| 63 | ALG1 | 2570112 | 17557 | -0.999 | 0.0571 | No |