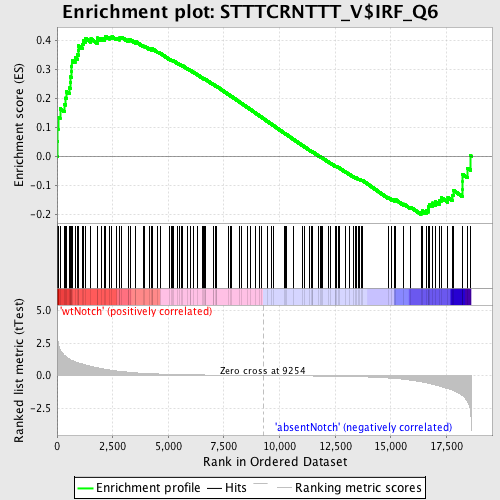
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #wtNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#wtNotch_versus_absentNotch |
| Upregulated in class | wtNotch |
| GeneSet | STTTCRNTTT_V$IRF_Q6 |
| Enrichment Score (ES) | 0.41356257 |
| Normalized Enrichment Score (NES) | 1.253697 |
| Nominal p-value | 0.03935185 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HIP1R | 3120368 | 13 | 3.030 | 0.0498 | Yes | ||
| 2 | PSMB10 | 630504 6510039 | 30 | 2.673 | 0.0935 | Yes | ||
| 3 | CCND1 | 460524 770309 3120576 6980398 | 45 | 2.532 | 0.1349 | Yes | ||
| 4 | PSMB9 | 6980471 | 135 | 2.011 | 0.1636 | Yes | ||
| 5 | RNF31 | 110035 | 332 | 1.543 | 0.1787 | Yes | ||
| 6 | SLC12A7 | 6860079 | 382 | 1.465 | 0.2005 | Yes | ||
| 7 | LGALS3BP | 840692 | 416 | 1.415 | 0.2222 | Yes | ||
| 8 | TAPBP | 1980110 | 561 | 1.248 | 0.2353 | Yes | ||
| 9 | TCIRG1 | 4590524 5910079 6900619 | 590 | 1.223 | 0.2541 | Yes | ||
| 10 | BAT5 | 5720687 | 610 | 1.206 | 0.2732 | Yes | ||
| 11 | PSME1 | 450193 4480035 | 643 | 1.183 | 0.2912 | Yes | ||
| 12 | GSDMDC1 | 4010092 5050091 | 653 | 1.174 | 0.3102 | Yes | ||
| 13 | PSME2 | 4850086 | 669 | 1.162 | 0.3288 | Yes | ||
| 14 | VAMP5 | 3830300 | 809 | 1.062 | 0.3390 | Yes | ||
| 15 | RPL23 | 2260452 4850435 6450673 | 907 | 0.999 | 0.3504 | Yes | ||
| 16 | ARHGEF6 | 4780164 6380450 | 950 | 0.984 | 0.3645 | Yes | ||
| 17 | IL12RB1 | 2230338 5420053 | 964 | 0.975 | 0.3800 | Yes | ||
| 18 | PIK3R3 | 1770333 | 1147 | 0.876 | 0.3848 | Yes | ||
| 19 | TAP1 | 4050047 | 1179 | 0.854 | 0.3973 | Yes | ||
| 20 | YWHAG | 3780341 | 1275 | 0.804 | 0.4056 | Yes | ||
| 21 | IFIT2 | 4810735 | 1521 | 0.703 | 0.4040 | Yes | ||
| 22 | MLLT3 | 510204 3850722 6200088 | 1801 | 0.598 | 0.3989 | Yes | ||
| 23 | SFMBT1 | 2360450 2370070 | 1827 | 0.586 | 0.4073 | Yes | ||
| 24 | ITGB7 | 4670619 | 2006 | 0.529 | 0.4065 | Yes | ||
| 25 | NFATC1 | 510400 2320348 4050600 6180161 6290136 6620086 | 2151 | 0.485 | 0.4068 | Yes | ||
| 26 | MOV10 | 50673 430577 3870280 | 2174 | 0.478 | 0.4136 | Yes | ||
| 27 | PSMB8 | 2850707 | 2362 | 0.424 | 0.4105 | No | ||
| 28 | BST2 | 6760487 | 2465 | 0.396 | 0.4116 | No | ||
| 29 | PPP1R10 | 110136 3170725 3610484 | 2675 | 0.349 | 0.4061 | No | ||
| 30 | SLC25A28 | 6590600 | 2788 | 0.325 | 0.4055 | No | ||
| 31 | EIF5A | 1500059 5290022 6370026 | 2822 | 0.319 | 0.4090 | No | ||
| 32 | DDX58 | 2450075 | 2915 | 0.303 | 0.4091 | No | ||
| 33 | CHGB | 1980053 | 3202 | 0.253 | 0.3978 | No | ||
| 34 | MRPS18B | 5570086 | 3225 | 0.249 | 0.4008 | No | ||
| 35 | PSMA5 | 5390537 | 3301 | 0.238 | 0.4007 | No | ||
| 36 | WNT5A | 840685 3120152 | 3512 | 0.211 | 0.3928 | No | ||
| 37 | BCL11A | 6860369 | 3523 | 0.209 | 0.3958 | No | ||
| 38 | PSIP1 | 1780082 3190435 5050594 | 3876 | 0.172 | 0.3796 | No | ||
| 39 | UBE1L | 6660242 | 3944 | 0.166 | 0.3787 | No | ||
| 40 | NT5C3 | 1570075 | 4154 | 0.147 | 0.3699 | No | ||
| 41 | TNFSF13B | 1940438 6510441 | 4253 | 0.140 | 0.3669 | No | ||
| 42 | MXI1 | 5050064 5130484 | 4270 | 0.139 | 0.3683 | No | ||
| 43 | DDR2 | 7050273 | 4279 | 0.138 | 0.3702 | No | ||
| 44 | HOXB6 | 4210739 | 4529 | 0.121 | 0.3588 | No | ||
| 45 | MAP3K11 | 7000039 | 4629 | 0.115 | 0.3553 | No | ||
| 46 | TRPM3 | 2470110 6400731 6550047 | 5041 | 0.093 | 0.3346 | No | ||
| 47 | IL17RC | 630164 2810593 | 5132 | 0.089 | 0.3312 | No | ||
| 48 | CREB1 | 1500717 2230358 3610600 6550601 | 5196 | 0.087 | 0.3293 | No | ||
| 49 | PBEF1 | 1090278 | 5250 | 0.084 | 0.3278 | No | ||
| 50 | OTX1 | 2510215 | 5422 | 0.077 | 0.3199 | No | ||
| 51 | USF1 | 4280156 4610114 6370113 | 5511 | 0.074 | 0.3163 | No | ||
| 52 | KLHL13 | 6590731 | 5607 | 0.071 | 0.3124 | No | ||
| 53 | SLC15A3 | 6650333 | 5656 | 0.069 | 0.3109 | No | ||
| 54 | PHF15 | 6550066 6860673 | 5863 | 0.062 | 0.3008 | No | ||
| 55 | EGFL6 | 3940181 | 5979 | 0.058 | 0.2955 | No | ||
| 56 | PITX2 | 870537 2690139 | 6007 | 0.058 | 0.2950 | No | ||
| 57 | HGF | 3360593 | 6118 | 0.055 | 0.2900 | No | ||
| 58 | KIRREL3 | 2630195 | 6298 | 0.050 | 0.2812 | No | ||
| 59 | UBD | 5570632 | 6533 | 0.045 | 0.2692 | No | ||
| 60 | SATB2 | 2470181 | 6601 | 0.043 | 0.2663 | No | ||
| 61 | CBX4 | 70332 | 6634 | 0.043 | 0.2653 | No | ||
| 62 | BCL2L1 | 1580452 4200152 5420484 | 6668 | 0.042 | 0.2642 | No | ||
| 63 | INDO | 2680390 | 6683 | 0.041 | 0.2641 | No | ||
| 64 | MSX1 | 2650309 | 7013 | 0.035 | 0.2469 | No | ||
| 65 | GATA6 | 1770010 | 7041 | 0.034 | 0.2460 | No | ||
| 66 | LGP2 | 4480338 | 7136 | 0.033 | 0.2415 | No | ||
| 67 | IL4 | 6020537 | 7159 | 0.032 | 0.2408 | No | ||
| 68 | GSC | 1580685 | 7180 | 0.031 | 0.2403 | No | ||
| 69 | MSC | 5080139 | 7713 | 0.023 | 0.2119 | No | ||
| 70 | ELK4 | 460494 940369 4480136 | 7780 | 0.021 | 0.2087 | No | ||
| 71 | BHLHB3 | 3450438 | 7842 | 0.020 | 0.2057 | No | ||
| 72 | GRAP2 | 7100441 1410647 | 8197 | 0.015 | 0.1868 | No | ||
| 73 | NPR3 | 1580239 | 8281 | 0.014 | 0.1825 | No | ||
| 74 | DLX4 | 50215 6650059 | 8579 | 0.009 | 0.1666 | No | ||
| 75 | LGI1 | 1850020 | 8714 | 0.007 | 0.1595 | No | ||
| 76 | ALDH1A1 | 6520706 | 8936 | 0.004 | 0.1476 | No | ||
| 77 | CLDN3 | 2570524 | 9092 | 0.002 | 0.1392 | No | ||
| 78 | CXCL10 | 2450408 | 9169 | 0.001 | 0.1352 | No | ||
| 79 | IGFBP5 | 2360592 | 9436 | -0.003 | 0.1208 | No | ||
| 80 | HOXA13 | 3120647 | 9634 | -0.005 | 0.1102 | No | ||
| 81 | ASPA | 3290400 | 9707 | -0.006 | 0.1064 | No | ||
| 82 | PPARGC1A | 4670040 | 10233 | -0.014 | 0.0783 | No | ||
| 83 | IL22RA1 | 5720603 | 10274 | -0.014 | 0.0763 | No | ||
| 84 | ZBP1 | 4210138 | 10308 | -0.015 | 0.0748 | No | ||
| 85 | IL27 | 1990324 | 10643 | -0.020 | 0.0571 | No | ||
| 86 | PSMA3 | 5900047 7040161 | 11049 | -0.026 | 0.0356 | No | ||
| 87 | FMR1 | 5050075 | 11131 | -0.028 | 0.0317 | No | ||
| 88 | CPT1A | 6350093 | 11365 | -0.031 | 0.0196 | No | ||
| 89 | ECM2 | 1470561 | 11447 | -0.033 | 0.0157 | No | ||
| 90 | PGM5 | 2340446 | 11487 | -0.034 | 0.0142 | No | ||
| 91 | GSH1 | 2760242 | 11494 | -0.034 | 0.0144 | No | ||
| 92 | NEDD4 | 460736 1190280 | 11741 | -0.039 | 0.0018 | No | ||
| 93 | EDIL3 | 1240039 | 11830 | -0.040 | -0.0023 | No | ||
| 94 | HPCAL1 | 1980129 2350348 | 11878 | -0.041 | -0.0042 | No | ||
| 95 | ARHGAP5 | 2510619 3360035 | 11933 | -0.042 | -0.0064 | No | ||
| 96 | MEIS1 | 1400575 | 12198 | -0.048 | -0.0199 | No | ||
| 97 | NCF1 | 3850091 | 12308 | -0.051 | -0.0249 | No | ||
| 98 | TCF15 | 1690400 | 12509 | -0.055 | -0.0348 | No | ||
| 99 | PLXNC1 | 5860315 | 12515 | -0.056 | -0.0342 | No | ||
| 100 | LIF | 1410373 2060129 3990338 | 12546 | -0.056 | -0.0349 | No | ||
| 101 | LRP2 | 1400204 3290706 | 12636 | -0.059 | -0.0387 | No | ||
| 102 | ESR1 | 4060372 5860193 | 12674 | -0.060 | -0.0397 | No | ||
| 103 | BCOR | 1660452 3940053 | 12970 | -0.069 | -0.0545 | No | ||
| 104 | IFNB1 | 1400142 | 13131 | -0.075 | -0.0619 | No | ||
| 105 | THRAP1 | 2470121 | 13309 | -0.081 | -0.0701 | No | ||
| 106 | MAB21L1 | 1450634 1580092 | 13433 | -0.086 | -0.0754 | No | ||
| 107 | OSR2 | 70711 | 13442 | -0.086 | -0.0744 | No | ||
| 108 | SOX5 | 2370576 2900167 3190128 5050528 | 13552 | -0.091 | -0.0788 | No | ||
| 109 | MGAT4A | 5690471 | 13579 | -0.092 | -0.0786 | No | ||
| 110 | NR3C2 | 2480324 6400093 | 13702 | -0.097 | -0.0836 | No | ||
| 111 | LRMP | 6290193 | 13724 | -0.098 | -0.0831 | No | ||
| 112 | NRG1 | 1050332 | 14898 | -0.187 | -0.1435 | No | ||
| 113 | E2F3 | 50162 460180 | 15049 | -0.203 | -0.1482 | No | ||
| 114 | KCNE4 | 2470102 | 15170 | -0.220 | -0.1510 | No | ||
| 115 | ADAM12 | 3390132 4070347 | 15196 | -0.224 | -0.1487 | No | ||
| 116 | CDKN2C | 5050750 5130148 | 15580 | -0.286 | -0.1646 | No | ||
| 117 | COL4A1 | 1740575 | 15894 | -0.355 | -0.1757 | No | ||
| 118 | TCF12 | 3610324 7000156 | 16359 | -0.484 | -0.1927 | No | ||
| 119 | CITED2 | 5670114 5130088 | 16426 | -0.502 | -0.1879 | No | ||
| 120 | COL4A2 | 2350619 | 16597 | -0.560 | -0.1878 | No | ||
| 121 | SFRS6 | 60224 | 16682 | -0.590 | -0.1825 | No | ||
| 122 | PIGR | 6520441 | 16693 | -0.594 | -0.1731 | No | ||
| 123 | CDK6 | 4920253 | 16728 | -0.610 | -0.1648 | No | ||
| 124 | ETV6 | 610524 | 16866 | -0.666 | -0.1611 | No | ||
| 125 | CASP1 | 2360546 | 17014 | -0.729 | -0.1569 | No | ||
| 126 | EVL | 1740113 | 17197 | -0.818 | -0.1531 | No | ||
| 127 | ZDHHC14 | 3780458 | 17262 | -0.846 | -0.1425 | No | ||
| 128 | B2M | 5080332 5130059 | 17565 | -1.000 | -0.1422 | No | ||
| 129 | NFIX | 2450152 | 17781 | -1.136 | -0.1349 | No | ||
| 130 | CASP7 | 4210156 | 17814 | -1.162 | -0.1173 | No | ||
| 131 | VGLL4 | 6860463 | 18208 | -1.524 | -0.1131 | No | ||
| 132 | CHCHD1 | 110706 | 18214 | -1.537 | -0.0878 | No | ||
| 133 | USP18 | 4010110 | 18219 | -1.542 | -0.0623 | No | ||
| 134 | IL18BP | 3450128 | 18451 | -2.018 | -0.0412 | No | ||
| 135 | PTK2 | 1780148 | 18588 | -3.004 | 0.0015 | No |

