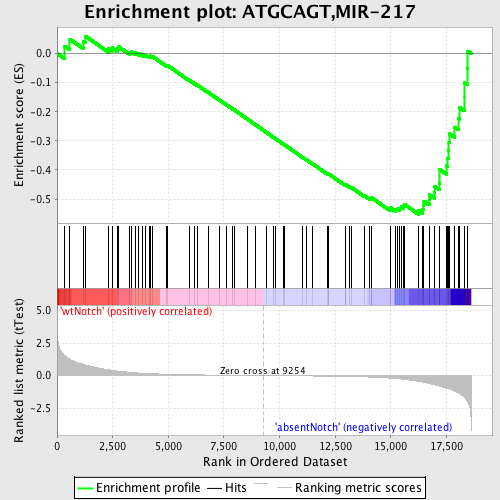
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #wtNotch_versus_absentNotch |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#wtNotch_versus_absentNotch |
| Upregulated in class | absentNotch |
| GeneSet | ATGCAGT,MIR-217 |
| Enrichment Score (ES) | -0.55298334 |
| Normalized Enrichment Score (NES) | -1.5148692 |
| Nominal p-value | 0.003690037 |
| FDR q-value | 0.1539676 |
| FWER p-Value | 0.847 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSMF1 | 5340056 | 344 | 1.521 | 0.0256 | No | ||
| 2 | OGT | 2360131 4610333 | 577 | 1.238 | 0.0491 | No | ||
| 3 | HIVEP3 | 3990551 | 1172 | 0.861 | 0.0420 | No | ||
| 4 | YWHAG | 3780341 | 1275 | 0.804 | 0.0599 | No | ||
| 5 | SLC31A1 | 4590161 6940154 | 2294 | 0.444 | 0.0179 | No | ||
| 6 | KCTD9 | 780577 | 2470 | 0.394 | 0.0199 | No | ||
| 7 | SLC39A10 | 4150347 | 2710 | 0.341 | 0.0169 | No | ||
| 8 | EHMT1 | 5290433 5910086 6620112 6980014 | 2760 | 0.332 | 0.0239 | No | ||
| 9 | STAG2 | 4540132 | 3252 | 0.245 | 0.0045 | No | ||
| 10 | ATP6V1A | 6590242 | 3325 | 0.234 | 0.0075 | No | ||
| 11 | BCL11A | 6860369 | 3523 | 0.209 | 0.0029 | No | ||
| 12 | KCNA3 | 510059 | 3678 | 0.191 | 0.0002 | No | ||
| 13 | MTPN | 3940167 | 3844 | 0.176 | -0.0036 | No | ||
| 14 | SLITRK3 | 5910041 | 3982 | 0.163 | -0.0063 | No | ||
| 15 | STX1A | 3060273 | 4143 | 0.148 | -0.0106 | No | ||
| 16 | NEUROD1 | 3060619 | 4179 | 0.145 | -0.0083 | No | ||
| 17 | AKAP2 | 5550347 | 4308 | 0.136 | -0.0112 | No | ||
| 18 | FN1 | 1170601 2970647 6220288 6940037 | 4923 | 0.099 | -0.0415 | No | ||
| 19 | GRIK2 | 4610164 | 4974 | 0.096 | -0.0414 | No | ||
| 20 | ST18 | 5720537 | 5940 | 0.060 | -0.0917 | No | ||
| 21 | ELOVL4 | 5220333 | 6188 | 0.053 | -0.1035 | No | ||
| 22 | ATP11C | 770672 7040129 | 6317 | 0.050 | -0.1090 | No | ||
| 23 | PAQR3 | 2480064 | 6793 | 0.039 | -0.1334 | No | ||
| 24 | STAT5B | 6200026 | 7297 | 0.029 | -0.1597 | No | ||
| 25 | DACH1 | 2450593 5700592 | 7633 | 0.024 | -0.1771 | No | ||
| 26 | GPM6A | 1660044 2750152 | 7864 | 0.020 | -0.1889 | No | ||
| 27 | FBXW11 | 6450632 | 7959 | 0.018 | -0.1935 | No | ||
| 28 | ACVR2A | 6110647 | 8573 | 0.009 | -0.2263 | No | ||
| 29 | ADAMTS5 | 5890592 | 8934 | 0.004 | -0.2456 | No | ||
| 30 | NFIA | 2760129 5860278 | 9392 | -0.002 | -0.2702 | No | ||
| 31 | TLK2 | 1170161 | 9725 | -0.007 | -0.2879 | No | ||
| 32 | STRBP | 4210594 5360239 | 9823 | -0.008 | -0.2929 | No | ||
| 33 | CHN2 | 870528 | 10171 | -0.013 | -0.3112 | No | ||
| 34 | DLGAP2 | 1090348 6840154 | 10232 | -0.014 | -0.3141 | No | ||
| 35 | GDI2 | 2370068 | 11033 | -0.026 | -0.3565 | No | ||
| 36 | TBC1D15 | 870673 | 11200 | -0.029 | -0.3646 | No | ||
| 37 | HNRPA2B1 | 1240619 6200161 6200494 6980408 | 11467 | -0.033 | -0.3780 | No | ||
| 38 | SEPT4 | 1580593 | 12137 | -0.047 | -0.4127 | No | ||
| 39 | TMSB4X | 6620114 | 12177 | -0.048 | -0.4134 | No | ||
| 40 | DMRT2 | 2630068 | 12183 | -0.048 | -0.4123 | No | ||
| 41 | SENP5 | 4560537 6760465 | 12954 | -0.069 | -0.4518 | No | ||
| 42 | MAMDC2 | 430471 | 12983 | -0.070 | -0.4513 | No | ||
| 43 | ZCCHC2 | 460592 | 13138 | -0.075 | -0.4575 | No | ||
| 44 | BAI3 | 940692 | 13246 | -0.078 | -0.4610 | No | ||
| 45 | CUL5 | 450142 | 13798 | -0.101 | -0.4878 | No | ||
| 46 | KLF12 | 1660095 4810288 5340546 6520286 | 14054 | -0.117 | -0.4981 | No | ||
| 47 | A2BP1 | 2370390 4590593 5550014 | 14121 | -0.120 | -0.4982 | No | ||
| 48 | HNRPUL1 | 2120008 4920408 5570180 6370609 6770403 | 14128 | -0.121 | -0.4950 | No | ||
| 49 | SOX11 | 610279 | 14967 | -0.194 | -0.5346 | No | ||
| 50 | RIN2 | 2450184 | 14973 | -0.194 | -0.5292 | No | ||
| 51 | DYRK1A | 3190181 | 15214 | -0.226 | -0.5356 | No | ||
| 52 | SIRT1 | 1190731 | 15314 | -0.239 | -0.5340 | No | ||
| 53 | JOSD3 | 1110543 | 15407 | -0.254 | -0.5316 | No | ||
| 54 | EZH2 | 6130605 6380524 | 15457 | -0.264 | -0.5266 | No | ||
| 55 | MSN | 4060044 | 15560 | -0.282 | -0.5239 | No | ||
| 56 | RAP2C | 1690132 | 15592 | -0.288 | -0.5172 | No | ||
| 57 | CAPN3 | 1980451 6940047 | 16257 | -0.449 | -0.5399 | Yes | ||
| 58 | FBN2 | 50435 | 16435 | -0.505 | -0.5348 | Yes | ||
| 59 | MYEF2 | 6860484 | 16475 | -0.517 | -0.5219 | Yes | ||
| 60 | MAPK1 | 3190193 6200253 | 16488 | -0.522 | -0.5074 | Yes | ||
| 61 | RTF1 | 2100735 | 16724 | -0.609 | -0.5024 | Yes | ||
| 62 | TACC2 | 580092 1400364 2650497 2690315 5360338 | 16737 | -0.614 | -0.4852 | Yes | ||
| 63 | KRAS | 2060170 | 16950 | -0.698 | -0.4764 | Yes | ||
| 64 | APRIN | 2810050 | 16956 | -0.700 | -0.4563 | Yes | ||
| 65 | NR4A2 | 60273 | 17174 | -0.804 | -0.4447 | Yes | ||
| 66 | ATP1B1 | 3130594 | 17202 | -0.820 | -0.4223 | Yes | ||
| 67 | PCNA | 940754 | 17203 | -0.821 | -0.3984 | Yes | ||
| 68 | POLG | 780465 | 17498 | -0.979 | -0.3859 | Yes | ||
| 69 | EIF4A2 | 1170494 1740711 2850504 | 17540 | -0.997 | -0.3591 | Yes | ||
| 70 | SLC38A2 | 1470242 3800026 | 17587 | -1.006 | -0.3324 | Yes | ||
| 71 | DYNC1LI2 | 50338 | 17612 | -1.019 | -0.3041 | Yes | ||
| 72 | YAF2 | 940053 | 17633 | -1.027 | -0.2754 | Yes | ||
| 73 | NDEL1 | 2370465 | 17875 | -1.204 | -0.2534 | Yes | ||
| 74 | UBL3 | 6450458 | 18048 | -1.341 | -0.2237 | Yes | ||
| 75 | IVNS1ABP | 4760601 6520113 | 18102 | -1.389 | -0.1863 | Yes | ||
| 76 | APPBP2 | 5130215 | 18309 | -1.671 | -0.1488 | Yes | ||
| 77 | ZFYVE20 | 6420400 | 18311 | -1.675 | -0.1003 | Yes | ||
| 78 | STX6 | 4850750 6770278 | 18434 | -1.963 | -0.0498 | Yes | ||
| 79 | PPM1D | 1690193 | 18460 | -2.052 | 0.0084 | Yes |

