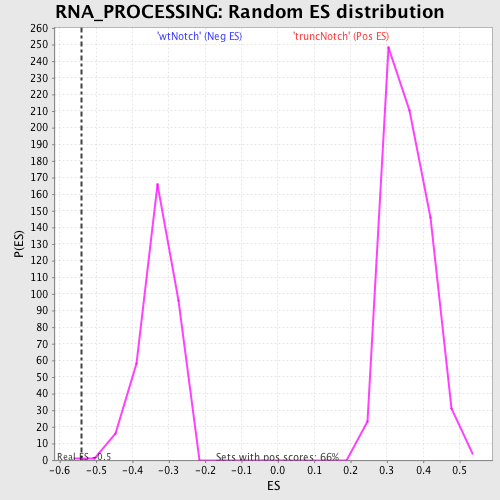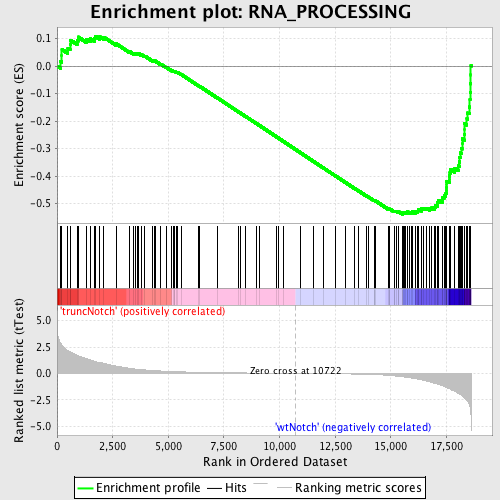
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | RNA_PROCESSING |
| Enrichment Score (ES) | -0.5398581 |
| Normalized Enrichment Score (NES) | -1.631286 |
| Nominal p-value | 0.00295858 |
| FDR q-value | 0.3012507 |
| FWER p-Value | 0.936 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SFRS2 | 50707 380593 | 159 | 2.828 | 0.0164 | No | ||
| 2 | IGF2BP3 | 2810161 4230338 | 214 | 2.664 | 0.0370 | No | ||
| 3 | GEMIN7 | 2320433 | 218 | 2.656 | 0.0602 | No | ||
| 4 | SF3A2 | 3140500 | 485 | 2.158 | 0.0649 | No | ||
| 5 | SNRPD2 | 6110035 | 586 | 2.033 | 0.0774 | No | ||
| 6 | METTL1 | 4540670 5860162 7040433 | 597 | 2.025 | 0.0948 | No | ||
| 7 | RBPMS | 3990494 | 900 | 1.713 | 0.0936 | No | ||
| 8 | SRRM1 | 6110025 | 948 | 1.674 | 0.1058 | No | ||
| 9 | NOLA2 | 4060167 | 1312 | 1.388 | 0.0984 | No | ||
| 10 | DDX54 | 6220324 | 1499 | 1.267 | 0.0996 | No | ||
| 11 | RRP9 | 630451 | 1671 | 1.161 | 0.1006 | No | ||
| 12 | SNRPB | 1230097 2940086 3520671 | 1704 | 1.142 | 0.1089 | No | ||
| 13 | LSM3 | 780164 | 1918 | 1.021 | 0.1064 | No | ||
| 14 | HNRPDL | 1050102 1090181 5360471 | 2100 | 0.946 | 0.1050 | No | ||
| 15 | NUDT21 | 1240167 | 2650 | 0.696 | 0.0814 | No | ||
| 16 | U2AF1 | 5130292 | 3252 | 0.480 | 0.0532 | No | ||
| 17 | LSM4 | 3130180 | 3437 | 0.430 | 0.0470 | No | ||
| 18 | SF3B4 | 110072 | 3511 | 0.414 | 0.0467 | No | ||
| 19 | TXNL4A | 4120286 6510242 | 3624 | 0.391 | 0.0441 | No | ||
| 20 | ELAVL4 | 50735 3360086 5220167 | 3674 | 0.381 | 0.0448 | No | ||
| 21 | SNRP70 | 4200181 | 3783 | 0.355 | 0.0421 | No | ||
| 22 | FARS2 | 5340575 | 3947 | 0.329 | 0.0362 | No | ||
| 23 | INTS6 | 6040736 7570767 | 4297 | 0.272 | 0.0198 | No | ||
| 24 | DHX16 | 940044 | 4372 | 0.263 | 0.0181 | No | ||
| 25 | HNRPUL1 | 2120008 4920408 5570180 6370609 6770403 | 4403 | 0.258 | 0.0187 | No | ||
| 26 | SFRS5 | 3450176 6350008 | 4650 | 0.228 | 0.0074 | No | ||
| 27 | BICD1 | 110128 2940324 | 4913 | 0.200 | -0.0050 | No | ||
| 28 | RBMY1A1 | 6200053 | 5156 | 0.177 | -0.0165 | No | ||
| 29 | RNPS1 | 610736 1090021 | 5235 | 0.170 | -0.0192 | No | ||
| 30 | SYNCRIP | 1690195 3140113 4670279 | 5278 | 0.167 | -0.0200 | No | ||
| 31 | CUGBP1 | 450292 510022 7050176 7050215 | 5294 | 0.166 | -0.0193 | No | ||
| 32 | PABPC4 | 1990170 6760270 5390138 | 5369 | 0.160 | -0.0219 | No | ||
| 33 | BCAS2 | 2340494 | 5393 | 0.158 | -0.0218 | No | ||
| 34 | NOL3 | 4060605 4670121 6620487 | 5578 | 0.145 | -0.0304 | No | ||
| 35 | NUFIP1 | 6350241 | 6343 | 0.102 | -0.0708 | No | ||
| 36 | FBL | 5130020 | 6411 | 0.099 | -0.0736 | No | ||
| 37 | SRPK1 | 450110 | 7205 | 0.070 | -0.1159 | No | ||
| 38 | RNGTT | 780152 780746 1090471 | 8169 | 0.045 | -0.1675 | No | ||
| 39 | EXOSC3 | 1740064 | 8245 | 0.044 | -0.1712 | No | ||
| 40 | SSB | 460286 3140717 | 8480 | 0.039 | -0.1835 | No | ||
| 41 | PRPF4B | 940181 2570300 | 8962 | 0.029 | -0.2092 | No | ||
| 42 | GEMIN6 | 2120288 3830176 | 9095 | 0.027 | -0.2161 | No | ||
| 43 | RBM5 | 3520402 5270121 | 9839 | 0.014 | -0.2562 | No | ||
| 44 | LSM5 | 4540398 | 9935 | 0.013 | -0.2612 | No | ||
| 45 | RBM6 | 840279 3840563 4780129 6020446 | 10181 | 0.009 | -0.2744 | No | ||
| 46 | HNRPF | 670138 | 10939 | -0.004 | -0.3153 | No | ||
| 47 | SNRPB2 | 5720577 | 11503 | -0.013 | -0.3456 | No | ||
| 48 | RPS14 | 430541 | 11990 | -0.022 | -0.3717 | No | ||
| 49 | CPSF3 | 2480671 | 12515 | -0.034 | -0.3997 | No | ||
| 50 | GEMIN5 | 3120577 | 12968 | -0.047 | -0.4237 | No | ||
| 51 | PRPF31 | 6100360 | 13350 | -0.061 | -0.4438 | No | ||
| 52 | SLBP | 6220601 | 13541 | -0.069 | -0.4535 | No | ||
| 53 | UPF1 | 430164 1190750 | 13888 | -0.087 | -0.4714 | No | ||
| 54 | PPARGC1A | 4670040 | 13995 | -0.094 | -0.4763 | No | ||
| 55 | SFRS9 | 3800047 | 14243 | -0.112 | -0.4887 | No | ||
| 56 | NONO | 7050014 | 14261 | -0.114 | -0.4886 | No | ||
| 57 | HNRPD | 4120021 | 14326 | -0.120 | -0.4910 | No | ||
| 58 | DHX8 | 6100139 | 14910 | -0.188 | -0.5208 | No | ||
| 59 | EXOSC2 | 6900471 | 14918 | -0.189 | -0.5196 | No | ||
| 60 | SNRPN | 4670195 5890494 6620110 | 14960 | -0.196 | -0.5200 | No | ||
| 61 | SNRPA1 | 1050735 6110131 | 15150 | -0.231 | -0.5282 | No | ||
| 62 | SFRS8 | 5130162 5700164 | 15177 | -0.235 | -0.5275 | No | ||
| 63 | NOL5A | 5570139 | 15237 | -0.250 | -0.5285 | No | ||
| 64 | PABPC1 | 2650180 2690253 6020632 1990270 | 15324 | -0.268 | -0.5308 | No | ||
| 65 | ZNF346 | 2650577 3990170 4150348 | 15351 | -0.273 | -0.5298 | No | ||
| 66 | PTBP1 | 4070736 | 15538 | -0.319 | -0.5370 | Yes | ||
| 67 | PRPF3 | 2510435 2940593 3610113 5390722 | 15542 | -0.320 | -0.5344 | Yes | ||
| 68 | ADAT1 | 510390 6900019 | 15571 | -0.326 | -0.5330 | Yes | ||
| 69 | PRPF8 | 1230497 1570600 | 15625 | -0.345 | -0.5328 | Yes | ||
| 70 | SNRPG | 2340551 | 15653 | -0.353 | -0.5312 | Yes | ||
| 71 | NOLC1 | 2350195 | 15741 | -0.381 | -0.5325 | Yes | ||
| 72 | SFRS10 | 6130528 6400438 7040112 | 15755 | -0.384 | -0.5298 | Yes | ||
| 73 | DDX39 | 3840148 | 15857 | -0.421 | -0.5316 | Yes | ||
| 74 | SNW1 | 4010736 | 15922 | -0.442 | -0.5311 | Yes | ||
| 75 | SIP1 | 2640706 | 15960 | -0.456 | -0.5291 | Yes | ||
| 76 | PPAN | 540398 | 16095 | -0.498 | -0.5320 | Yes | ||
| 77 | RBM3 | 2360739 5700167 | 16115 | -0.506 | -0.5285 | Yes | ||
| 78 | EXOSC7 | 520538 | 16197 | -0.537 | -0.5282 | Yes | ||
| 79 | DUSP11 | 1500088 4610280 | 16225 | -0.548 | -0.5248 | Yes | ||
| 80 | EFTUD2 | 2470707 4610148 | 16247 | -0.554 | -0.5210 | Yes | ||
| 81 | FUSIP1 | 520082 5390114 | 16369 | -0.602 | -0.5223 | Yes | ||
| 82 | CRNKL1 | 5050097 | 16380 | -0.607 | -0.5174 | Yes | ||
| 83 | SNRPD1 | 4480162 | 16482 | -0.653 | -0.5171 | Yes | ||
| 84 | SNRPF | 3170280 | 16586 | -0.700 | -0.5165 | Yes | ||
| 85 | CSTF1 | 4610129 | 16731 | -0.795 | -0.5173 | Yes | ||
| 86 | USP39 | 1090242 3520435 4760736 | 16845 | -0.862 | -0.5158 | Yes | ||
| 87 | RBMS1 | 6400014 | 16951 | -0.929 | -0.5133 | Yes | ||
| 88 | SF3A1 | 1580315 | 17005 | -0.970 | -0.5076 | Yes | ||
| 89 | GEMIN4 | 130278 2230075 | 17092 | -1.003 | -0.5034 | Yes | ||
| 90 | SNRPD3 | 2970050 | 17096 | -1.004 | -0.4947 | Yes | ||
| 91 | DGCR8 | 3450632 | 17135 | -1.022 | -0.4877 | Yes | ||
| 92 | SF3B2 | 6590576 | 17310 | -1.148 | -0.4870 | Yes | ||
| 93 | KHDRBS1 | 1240403 6040040 | 17319 | -1.152 | -0.4772 | Yes | ||
| 94 | SFRS7 | 2760408 | 17394 | -1.224 | -0.4704 | Yes | ||
| 95 | PHF5A | 2690519 | 17471 | -1.284 | -0.4632 | Yes | ||
| 96 | CPSF1 | 6290064 | 17492 | -1.314 | -0.4527 | Yes | ||
| 97 | SFRS2IP | 730687 1500373 | 17499 | -1.317 | -0.4414 | Yes | ||
| 98 | RNMT | 1770500 | 17507 | -1.327 | -0.4301 | Yes | ||
| 99 | ASCC3L1 | 6620494 | 17510 | -1.328 | -0.4184 | Yes | ||
| 100 | NCBP2 | 5340500 | 17626 | -1.442 | -0.4119 | Yes | ||
| 101 | SF3B3 | 3710044 | 17631 | -1.449 | -0.3993 | Yes | ||
| 102 | DHX38 | 780243 | 17656 | -1.474 | -0.3876 | Yes | ||
| 103 | TRNT1 | 870053 | 17689 | -1.498 | -0.3761 | Yes | ||
| 104 | SPOP | 450035 | 17876 | -1.710 | -0.3711 | Yes | ||
| 105 | SRPK2 | 6380341 | 18036 | -1.872 | -0.3632 | Yes | ||
| 106 | AARS | 1570279 2370136 | 18075 | -1.916 | -0.3483 | Yes | ||
| 107 | NSUN2 | 2120193 | 18099 | -1.961 | -0.3323 | Yes | ||
| 108 | ZNF638 | 6510112 | 18122 | -1.998 | -0.3158 | Yes | ||
| 109 | DHX15 | 870632 | 18188 | -2.087 | -0.3009 | Yes | ||
| 110 | IVNS1ABP | 4760601 6520113 | 18205 | -2.117 | -0.2831 | Yes | ||
| 111 | POP4 | 4560600 | 18210 | -2.122 | -0.2646 | Yes | ||
| 112 | SF3A3 | 6660603 | 18303 | -2.281 | -0.2494 | Yes | ||
| 113 | CDC2L2 | 1400671 5290168 | 18313 | -2.315 | -0.2294 | Yes | ||
| 114 | KIN | 6550014 | 18316 | -2.324 | -0.2090 | Yes | ||
| 115 | GRSF1 | 2100184 | 18421 | -2.543 | -0.1922 | Yes | ||
| 116 | DDX20 | 3290348 | 18454 | -2.649 | -0.1706 | Yes | ||
| 117 | CSTF3 | 3850156 5570458 | 18524 | -2.902 | -0.1487 | Yes | ||
| 118 | SFPQ | 4760110 | 18556 | -3.132 | -0.1227 | Yes | ||
| 119 | DBR1 | 1770286 | 18564 | -3.218 | -0.0947 | Yes | ||
| 120 | CSTF2 | 6040463 | 18583 | -3.529 | -0.0645 | Yes | ||
| 121 | SFRS6 | 60224 | 18598 | -3.719 | -0.0324 | Yes | ||
| 122 | SFRS1 | 2360440 | 18601 | -3.775 | 0.0008 | Yes |