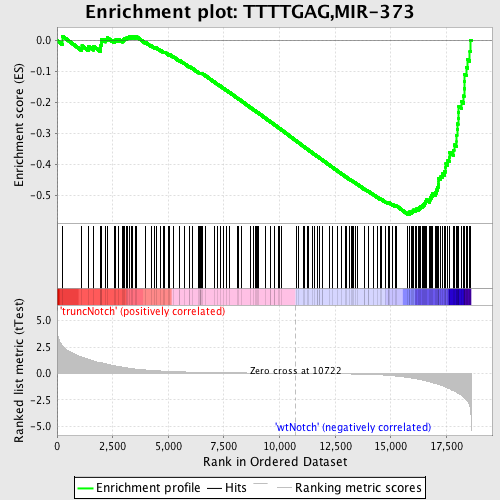
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | TTTTGAG,MIR-373 |
| Enrichment Score (ES) | -0.561729 |
| Normalized Enrichment Score (NES) | -1.7763475 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0060550836 |
| FWER p-Value | 0.033 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PRR7 | 6250291 | 240 | 2.589 | 0.0143 | No | ||
| 2 | RELB | 1400048 | 1117 | 1.525 | -0.0171 | No | ||
| 3 | FKBP4 | 870035 | 1414 | 1.330 | -0.0191 | No | ||
| 4 | PITPNM2 | 1580044 | 1630 | 1.183 | -0.0183 | No | ||
| 5 | BLNK | 70288 6590092 | 1942 | 1.009 | -0.0245 | No | ||
| 6 | CSK | 6350593 | 1964 | 1.003 | -0.0151 | No | ||
| 7 | EPHB2 | 1090288 1410377 2100541 | 1989 | 0.997 | -0.0058 | No | ||
| 8 | SLC38A2 | 1470242 3800026 | 2000 | 0.995 | 0.0041 | No | ||
| 9 | TARDBP | 870390 2350093 | 2164 | 0.917 | 0.0049 | No | ||
| 10 | CHST2 | 3180682 | 2249 | 0.876 | 0.0096 | No | ||
| 11 | PPP4C | 5130593 | 2571 | 0.736 | -0.0000 | No | ||
| 12 | OPRK1 | 3780632 | 2642 | 0.700 | 0.0036 | No | ||
| 13 | FRMD4A | 2190176 | 2763 | 0.644 | 0.0039 | No | ||
| 14 | ZFAND3 | 2760504 | 2926 | 0.591 | 0.0013 | No | ||
| 15 | KCTD12 | 7050091 | 2987 | 0.569 | 0.0041 | No | ||
| 16 | CFL2 | 380239 6200368 | 3035 | 0.551 | 0.0073 | No | ||
| 17 | ADRA1A | 1230446 2260390 | 3118 | 0.521 | 0.0084 | No | ||
| 18 | SOX9 | 5720681 | 3182 | 0.499 | 0.0102 | No | ||
| 19 | KPNA4 | 510369 | 3238 | 0.482 | 0.0123 | No | ||
| 20 | SYT4 | 5080193 | 3357 | 0.448 | 0.0107 | No | ||
| 21 | BUB3 | 3170546 | 3396 | 0.440 | 0.0132 | No | ||
| 22 | NAV1 | 4210519 6620129 | 3510 | 0.414 | 0.0115 | No | ||
| 23 | FBXW7 | 4210338 7050280 | 3558 | 0.405 | 0.0132 | No | ||
| 24 | ARMC8 | 520408 940132 4060368 4210102 | 3972 | 0.325 | -0.0058 | No | ||
| 25 | SH3GLB1 | 4590079 5670187 | 4231 | 0.279 | -0.0168 | No | ||
| 26 | FLRT2 | 4200035 | 4379 | 0.262 | -0.0220 | No | ||
| 27 | UBAP1 | 4280059 | 4454 | 0.251 | -0.0234 | No | ||
| 28 | SFRS5 | 3450176 6350008 | 4650 | 0.228 | -0.0315 | No | ||
| 29 | PPP2R5C | 3310121 6650672 | 4798 | 0.212 | -0.0373 | No | ||
| 30 | SEH1L | 4590563 | 4839 | 0.208 | -0.0372 | No | ||
| 31 | FOXE1 | 4810164 | 5027 | 0.189 | -0.0454 | No | ||
| 32 | MTMR4 | 4540524 | 5056 | 0.186 | -0.0449 | No | ||
| 33 | RNPS1 | 610736 1090021 | 5235 | 0.170 | -0.0528 | No | ||
| 34 | ST3GAL5 | 2510446 | 5491 | 0.151 | -0.0650 | No | ||
| 35 | ARID5B | 110403 | 5496 | 0.151 | -0.0636 | No | ||
| 36 | PKNOX2 | 610066 2810075 4230601 | 5706 | 0.136 | -0.0735 | No | ||
| 37 | FBLN5 | 6550010 | 5965 | 0.121 | -0.0862 | No | ||
| 38 | HOXA3 | 3610632 6020358 | 5970 | 0.121 | -0.0852 | No | ||
| 39 | TMEPAI | 1340253 | 6077 | 0.115 | -0.0897 | No | ||
| 40 | HOXB5 | 2900538 | 6359 | 0.101 | -0.1038 | No | ||
| 41 | KDELR1 | 6760504 | 6416 | 0.099 | -0.1058 | No | ||
| 42 | CACNA1G | 3060161 4810706 5550722 | 6438 | 0.098 | -0.1059 | No | ||
| 43 | LGP2 | 4480338 | 6445 | 0.097 | -0.1052 | No | ||
| 44 | ADRBK2 | 1660692 | 6458 | 0.097 | -0.1049 | No | ||
| 45 | GRM3 | 4540242 | 6503 | 0.095 | -0.1063 | No | ||
| 46 | CLEC5A | 6420139 | 6516 | 0.095 | -0.1059 | No | ||
| 47 | PTPN5 | 2350161 | 6685 | 0.088 | -0.1141 | No | ||
| 48 | WAC | 5340390 5910164 | 7054 | 0.075 | -0.1332 | No | ||
| 49 | PAFAH1B1 | 4230333 6420121 6450066 | 7219 | 0.070 | -0.1414 | No | ||
| 50 | CAPN3 | 1980451 6940047 | 7325 | 0.067 | -0.1464 | No | ||
| 51 | MYO5C | 3850097 | 7474 | 0.062 | -0.1537 | No | ||
| 52 | FBXO28 | 1340358 4730100 | 7623 | 0.059 | -0.1611 | No | ||
| 53 | TMEFF1 | 2680100 | 7762 | 0.055 | -0.1680 | No | ||
| 54 | SUHW4 | 7000075 | 8104 | 0.047 | -0.1860 | No | ||
| 55 | RNGTT | 780152 780746 1090471 | 8169 | 0.045 | -0.1890 | No | ||
| 56 | MS4A2 | 2940594 | 8301 | 0.042 | -0.1957 | No | ||
| 57 | GABRB3 | 4150164 | 8713 | 0.034 | -0.2176 | No | ||
| 58 | HIF1A | 5670605 | 8830 | 0.032 | -0.2235 | No | ||
| 59 | ELOVL2 | 5720064 | 8906 | 0.030 | -0.2273 | No | ||
| 60 | RAP1A | 1090025 | 8954 | 0.029 | -0.2295 | No | ||
| 61 | ALK | 730008 | 9014 | 0.028 | -0.2324 | No | ||
| 62 | ADAMDEC1 | 3990110 | 9043 | 0.028 | -0.2336 | No | ||
| 63 | ZCCHC14 | 5900288 | 9387 | 0.022 | -0.2520 | No | ||
| 64 | SERTAD2 | 2810673 6840450 | 9604 | 0.019 | -0.2635 | No | ||
| 65 | TEX12 | 1090110 | 9612 | 0.018 | -0.2637 | No | ||
| 66 | MYB | 1660494 5860451 6130706 | 9771 | 0.015 | -0.2721 | No | ||
| 67 | MPP5 | 2100148 5220632 | 9784 | 0.015 | -0.2726 | No | ||
| 68 | ARHGAP5 | 2510619 3360035 | 9961 | 0.013 | -0.2820 | No | ||
| 69 | LENG8 | 3130731 | 10009 | 0.012 | -0.2844 | No | ||
| 70 | ISL1 | 4810239 | 10082 | 0.010 | -0.2882 | No | ||
| 71 | SLC4A4 | 1780075 1850040 4810068 | 10740 | -0.000 | -0.3238 | No | ||
| 72 | XPO1 | 540707 | 10863 | -0.002 | -0.3304 | No | ||
| 73 | KHDRBS2 | 3520717 | 11079 | -0.006 | -0.3420 | No | ||
| 74 | MAFB | 1230471 | 11103 | -0.006 | -0.3432 | No | ||
| 75 | KLF5 | 3840348 | 11248 | -0.009 | -0.3509 | No | ||
| 76 | ALDH18A1 | 2340601 | 11293 | -0.009 | -0.3532 | No | ||
| 77 | TRHDE | 1690068 | 11484 | -0.012 | -0.3633 | No | ||
| 78 | SSX2IP | 4610176 | 11580 | -0.014 | -0.3683 | No | ||
| 79 | NDFIP1 | 1980402 | 11696 | -0.017 | -0.3744 | No | ||
| 80 | PLAG1 | 1450142 3870139 6520039 | 11711 | -0.017 | -0.3749 | No | ||
| 81 | MAMDC1 | 3710129 4480075 | 11794 | -0.018 | -0.3792 | No | ||
| 82 | SLC17A7 | 7000037 | 11806 | -0.019 | -0.3796 | No | ||
| 83 | CCNJ | 870047 | 11944 | -0.021 | -0.3868 | No | ||
| 84 | ZIC2 | 1410408 | 11949 | -0.021 | -0.3868 | No | ||
| 85 | GNAI1 | 4560390 | 12262 | -0.028 | -0.4034 | No | ||
| 86 | KCNIP2 | 60088 1780324 | 12386 | -0.031 | -0.4097 | No | ||
| 87 | NF2 | 4150735 6450139 | 12588 | -0.036 | -0.4202 | No | ||
| 88 | GOLGA4 | 460300 | 12780 | -0.042 | -0.4301 | No | ||
| 89 | DSCAML1 | 1850403 5550079 | 12978 | -0.047 | -0.4403 | No | ||
| 90 | WTAP | 4010176 | 12992 | -0.048 | -0.4405 | No | ||
| 91 | PTP4A1 | 130692 | 13161 | -0.054 | -0.4491 | No | ||
| 92 | ITGA10 | 6330438 | 13221 | -0.056 | -0.4517 | No | ||
| 93 | KLHL13 | 6590731 | 13297 | -0.059 | -0.4551 | No | ||
| 94 | SP2 | 6400026 | 13304 | -0.060 | -0.4548 | No | ||
| 95 | ACTR2 | 1170332 | 13428 | -0.064 | -0.4608 | No | ||
| 96 | FGFR2 | 4670601 5570402 6940377 | 13521 | -0.069 | -0.4650 | No | ||
| 97 | EN2 | 7000368 | 13806 | -0.082 | -0.4796 | No | ||
| 98 | DNAJB6 | 2810309 5570451 | 13819 | -0.083 | -0.4793 | No | ||
| 99 | PPARGC1A | 4670040 | 13995 | -0.094 | -0.4878 | No | ||
| 100 | CAPZA2 | 110427 4590064 | 14001 | -0.094 | -0.4871 | No | ||
| 101 | PALM2 | 4120068 7100398 | 14206 | -0.109 | -0.4970 | No | ||
| 102 | ARF4 | 3190661 | 14414 | -0.127 | -0.5069 | No | ||
| 103 | KPNA3 | 7040088 | 14523 | -0.137 | -0.5113 | No | ||
| 104 | ORC2L | 1990470 6510019 | 14587 | -0.144 | -0.5132 | No | ||
| 105 | GOPC | 6620400 | 14775 | -0.168 | -0.5216 | No | ||
| 106 | JAK2 | 3780528 6100692 6550577 | 14873 | -0.182 | -0.5249 | No | ||
| 107 | ARID4A | 520156 1050707 | 14880 | -0.183 | -0.5233 | No | ||
| 108 | RNF12 | 6860717 | 14931 | -0.191 | -0.5240 | No | ||
| 109 | CLASP1 | 6860279 | 15087 | -0.218 | -0.5301 | No | ||
| 110 | PPP3CA | 4760332 6760092 | 15201 | -0.243 | -0.5337 | No | ||
| 111 | WDFY1 | 1580463 | 15257 | -0.253 | -0.5340 | No | ||
| 112 | SFRS10 | 6130528 6400438 7040112 | 15755 | -0.384 | -0.5569 | No | ||
| 113 | MBP | 5220369 | 15846 | -0.417 | -0.5573 | Yes | ||
| 114 | YTHDF3 | 4540632 4670717 7040609 | 15856 | -0.421 | -0.5534 | Yes | ||
| 115 | MOBKL2B | 940128 | 15949 | -0.452 | -0.5536 | Yes | ||
| 116 | UBE3A | 1240152 2690438 5860609 | 15982 | -0.461 | -0.5505 | Yes | ||
| 117 | PKP4 | 3710400 5890097 | 16012 | -0.470 | -0.5471 | Yes | ||
| 118 | FNBP4 | 1740139 7050113 | 16114 | -0.506 | -0.5472 | Yes | ||
| 119 | FN1 | 1170601 2970647 6220288 6940037 | 16151 | -0.517 | -0.5437 | Yes | ||
| 120 | ING4 | 510632 2340619 2850176 5290706 | 16248 | -0.554 | -0.5431 | Yes | ||
| 121 | ELOVL5 | 3800170 | 16300 | -0.577 | -0.5398 | Yes | ||
| 122 | ARFGEF1 | 6760494 | 16345 | -0.593 | -0.5359 | Yes | ||
| 123 | WDR44 | 130195 1050609 1570685 | 16412 | -0.621 | -0.5329 | Yes | ||
| 124 | VPS26A | 1940494 5720059 | 16455 | -0.639 | -0.5285 | Yes | ||
| 125 | DAG1 | 460053 610341 | 16520 | -0.673 | -0.5248 | Yes | ||
| 126 | CSNK1E | 2850347 5050093 6110301 | 16546 | -0.684 | -0.5190 | Yes | ||
| 127 | PURB | 5360138 | 16587 | -0.701 | -0.5137 | Yes | ||
| 128 | CAMK4 | 1690091 | 16754 | -0.806 | -0.5142 | Yes | ||
| 129 | ZC3H6 | 130711 | 16771 | -0.817 | -0.5065 | Yes | ||
| 130 | KRIT1 | 2640500 4230435 4730184 | 16823 | -0.851 | -0.5003 | Yes | ||
| 131 | TRIM33 | 580619 2230280 3990433 6200747 | 16870 | -0.879 | -0.4935 | Yes | ||
| 132 | DCUN1D1 | 2360332 | 17007 | -0.972 | -0.4906 | Yes | ||
| 133 | PSIP1 | 1780082 3190435 5050594 | 17048 | -0.987 | -0.4824 | Yes | ||
| 134 | TNRC6A | 1240537 6130037 6200010 | 17091 | -1.002 | -0.4741 | Yes | ||
| 135 | USP7 | 3990356 | 17128 | -1.018 | -0.4653 | Yes | ||
| 136 | EI24 | 3120433 | 17145 | -1.027 | -0.4554 | Yes | ||
| 137 | CACNG4 | 6980112 | 17149 | -1.031 | -0.4447 | Yes | ||
| 138 | PTER | 3450685 | 17251 | -1.102 | -0.4385 | Yes | ||
| 139 | CASP8 | 4850154 | 17331 | -1.166 | -0.4305 | Yes | ||
| 140 | RAB11A | 5360079 | 17419 | -1.242 | -0.4221 | Yes | ||
| 141 | RANBP5 | 3780019 | 17469 | -1.283 | -0.4112 | Yes | ||
| 142 | SOX4 | 2260091 | 17476 | -1.291 | -0.3980 | Yes | ||
| 143 | SLC23A2 | 6660722 | 17562 | -1.380 | -0.3880 | Yes | ||
| 144 | HDAC1 | 2850670 | 17640 | -1.456 | -0.3768 | Yes | ||
| 145 | EIF4A1 | 1990341 2810300 | 17648 | -1.463 | -0.3618 | Yes | ||
| 146 | PHTF2 | 6590053 | 17821 | -1.656 | -0.3536 | Yes | ||
| 147 | TOB1 | 4150138 | 17841 | -1.671 | -0.3370 | Yes | ||
| 148 | SENP1 | 6100537 | 17960 | -1.792 | -0.3246 | Yes | ||
| 149 | AP1G1 | 2360671 | 17963 | -1.793 | -0.3058 | Yes | ||
| 150 | SURF4 | 6020315 | 17975 | -1.816 | -0.2872 | Yes | ||
| 151 | AMMECR1 | 3800435 | 18003 | -1.846 | -0.2692 | Yes | ||
| 152 | PTPLB | 4810435 | 18022 | -1.863 | -0.2505 | Yes | ||
| 153 | VCP | 5340168 | 18033 | -1.869 | -0.2314 | Yes | ||
| 154 | EEF1A1 | 1980193 1990195 4670735 | 18057 | -1.897 | -0.2126 | Yes | ||
| 155 | MBNL2 | 3450707 | 18177 | -2.076 | -0.1972 | Yes | ||
| 156 | PABPN1 | 460519 | 18274 | -2.228 | -0.1789 | Yes | ||
| 157 | RANBP10 | 2340184 | 18294 | -2.269 | -0.1560 | Yes | ||
| 158 | ARFIP1 | 3130601 | 18306 | -2.289 | -0.1324 | Yes | ||
| 159 | PELI1 | 3870215 6900040 | 18317 | -2.326 | -0.1085 | Yes | ||
| 160 | BRD2 | 1450300 5340484 6100605 | 18401 | -2.514 | -0.0865 | Yes | ||
| 161 | ARHGAP21 | 2100168 | 18455 | -2.649 | -0.0614 | Yes | ||
| 162 | UBE2G1 | 2230546 5420528 7040463 | 18555 | -3.099 | -0.0341 | Yes | ||
| 163 | CBFB | 1850551 2650035 | 18587 | -3.540 | 0.0016 | Yes |

