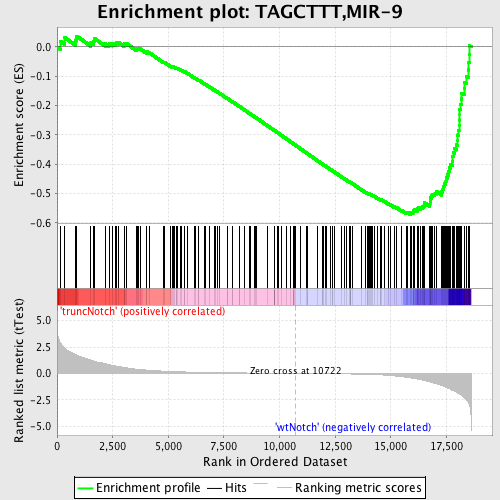
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | TAGCTTT,MIR-9 |
| Enrichment Score (ES) | -0.57116246 |
| Normalized Enrichment Score (NES) | -1.7882063 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.006049334 |
| FWER p-Value | 0.027 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SFRS2 | 50707 380593 | 159 | 2.828 | 0.0188 | No | ||
| 2 | HRB | 1660397 6110398 | 341 | 2.382 | 0.0321 | No | ||
| 3 | DTX3 | 1690458 | 816 | 1.796 | 0.0239 | No | ||
| 4 | TPM1 | 130673 | 883 | 1.728 | 0.0371 | No | ||
| 5 | ZMYND11 | 630181 2570019 4850138 5360040 | 1519 | 1.258 | 0.0149 | No | ||
| 6 | PITPNM2 | 1580044 | 1630 | 1.183 | 0.0204 | No | ||
| 7 | HNRPA2B1 | 1240619 6200161 6200494 6980408 | 1667 | 1.163 | 0.0297 | No | ||
| 8 | TARDBP | 870390 2350093 | 2164 | 0.917 | 0.0117 | No | ||
| 9 | SERF2 | 5720010 | 2333 | 0.840 | 0.0108 | No | ||
| 10 | CPEB2 | 4760338 | 2471 | 0.773 | 0.0109 | No | ||
| 11 | NCKIPSD | 2940156 | 2607 | 0.717 | 0.0105 | No | ||
| 12 | CAMK2G | 630097 | 2653 | 0.695 | 0.0148 | No | ||
| 13 | CYR61 | 1240408 5290026 4120452 6550008 | 2750 | 0.650 | 0.0159 | No | ||
| 14 | GPHN | 3290195 3840121 | 3009 | 0.562 | 0.0074 | No | ||
| 15 | CFL2 | 380239 6200368 | 3035 | 0.551 | 0.0114 | No | ||
| 16 | LRP12 | 2370112 | 3131 | 0.517 | 0.0113 | No | ||
| 17 | ATXN1 | 5550156 | 3574 | 0.402 | -0.0088 | No | ||
| 18 | PPP1R8 | 6290292 | 3591 | 0.397 | -0.0058 | No | ||
| 19 | GSPT2 | 6590674 | 3657 | 0.384 | -0.0056 | No | ||
| 20 | SLC9A9 | 1940309 | 3734 | 0.366 | -0.0062 | No | ||
| 21 | MAGI1 | 6510500 1990239 4670471 | 4023 | 0.316 | -0.0187 | No | ||
| 22 | AFF3 | 3870102 | 4032 | 0.315 | -0.0161 | No | ||
| 23 | PHF6 | 3120632 3990041 | 4159 | 0.291 | -0.0201 | No | ||
| 24 | SLITRK6 | 7320739 | 4786 | 0.213 | -0.0520 | No | ||
| 25 | HOXB13 | 4730152 | 4814 | 0.210 | -0.0514 | No | ||
| 26 | POU3F2 | 6620484 | 5113 | 0.181 | -0.0658 | No | ||
| 27 | BACH2 | 6760131 | 5165 | 0.177 | -0.0669 | No | ||
| 28 | HTR2C | 380497 | 5216 | 0.172 | -0.0679 | No | ||
| 29 | THBD | 1500092 | 5281 | 0.167 | -0.0697 | No | ||
| 30 | SOCS4 | 1170162 | 5375 | 0.160 | -0.0732 | No | ||
| 31 | SOX2 | 630487 2230491 | 5382 | 0.159 | -0.0720 | No | ||
| 32 | PI15 | 3840133 | 5423 | 0.155 | -0.0727 | No | ||
| 33 | TPD52 | 670463 4610541 | 5528 | 0.149 | -0.0769 | No | ||
| 34 | CREB5 | 2320368 | 5586 | 0.144 | -0.0786 | No | ||
| 35 | STX16 | 70315 | 5708 | 0.136 | -0.0838 | No | ||
| 36 | SOX21 | 1050110 4670041 6660338 | 5747 | 0.134 | -0.0846 | No | ||
| 37 | ELAVL2 | 360181 | 5855 | 0.127 | -0.0891 | No | ||
| 38 | PIGF | 5690050 | 6192 | 0.109 | -0.1063 | No | ||
| 39 | POU4F1 | 2260315 | 6227 | 0.108 | -0.1071 | No | ||
| 40 | HOXB5 | 2900538 | 6359 | 0.101 | -0.1132 | No | ||
| 41 | SOX3 | 4570537 | 6370 | 0.101 | -0.1128 | No | ||
| 42 | SLC16A1 | 50433 | 6612 | 0.091 | -0.1250 | No | ||
| 43 | PLCG1 | 6020369 | 6656 | 0.089 | -0.1264 | No | ||
| 44 | ZFPM2 | 460072 | 6833 | 0.082 | -0.1352 | No | ||
| 45 | WAC | 5340390 5910164 | 7054 | 0.075 | -0.1464 | No | ||
| 46 | OSBPL8 | 50603 | 7110 | 0.073 | -0.1486 | No | ||
| 47 | CALD1 | 1770129 1940397 | 7213 | 0.070 | -0.1535 | No | ||
| 48 | MEF2C | 670025 780338 | 7314 | 0.067 | -0.1583 | No | ||
| 49 | CPEB3 | 3940164 | 7648 | 0.058 | -0.1758 | No | ||
| 50 | NAP1L5 | 360576 | 7661 | 0.057 | -0.1759 | No | ||
| 51 | ENAH | 1690292 5700300 | 7869 | 0.052 | -0.1866 | No | ||
| 52 | LRRTM1 | 1690324 | 7886 | 0.052 | -0.1869 | No | ||
| 53 | PARD6B | 4120600 | 8185 | 0.045 | -0.2027 | No | ||
| 54 | FBXO22 | 4760010 | 8204 | 0.045 | -0.2032 | No | ||
| 55 | GPM6A | 1660044 2750152 | 8418 | 0.040 | -0.2144 | No | ||
| 56 | PRKCE | 5700053 | 8661 | 0.035 | -0.2271 | No | ||
| 57 | GABRB3 | 4150164 | 8713 | 0.034 | -0.2296 | No | ||
| 58 | CRISPLD1 | 5910524 | 8892 | 0.030 | -0.2389 | No | ||
| 59 | EPHA7 | 1190288 4010278 | 8903 | 0.030 | -0.2392 | No | ||
| 60 | PRPF4B | 940181 2570300 | 8962 | 0.029 | -0.2420 | No | ||
| 61 | SET | 6650286 | 9446 | 0.021 | -0.2680 | No | ||
| 62 | TEAD1 | 2470551 | 9750 | 0.016 | -0.2843 | No | ||
| 63 | DCX | 130075 5360025 7050673 | 9754 | 0.016 | -0.2843 | No | ||
| 64 | DPYSL3 | 1580594 | 9755 | 0.016 | -0.2841 | No | ||
| 65 | MPP5 | 2100148 5220632 | 9784 | 0.015 | -0.2855 | No | ||
| 66 | CCND2 | 5340167 | 9898 | 0.014 | -0.2915 | No | ||
| 67 | ARHGAP5 | 2510619 3360035 | 9961 | 0.013 | -0.2947 | No | ||
| 68 | ZFHX4 | 6110167 | 10096 | 0.010 | -0.3019 | No | ||
| 69 | TMEM47 | 1660086 | 10300 | 0.007 | -0.3129 | No | ||
| 70 | SMARCA1 | 4230315 | 10479 | 0.004 | -0.3225 | No | ||
| 71 | EDG1 | 4200619 | 10622 | 0.002 | -0.3302 | No | ||
| 72 | SP5 | 3840309 | 10628 | 0.002 | -0.3304 | No | ||
| 73 | AKT3 | 1580270 3290278 | 10642 | 0.001 | -0.3311 | No | ||
| 74 | ZFP36L1 | 2510138 4120048 | 10657 | 0.001 | -0.3318 | No | ||
| 75 | NDST1 | 1500427 5340121 | 10735 | -0.000 | -0.3360 | No | ||
| 76 | TLL2 | 6220040 | 10955 | -0.004 | -0.3479 | No | ||
| 77 | PCDH18 | 430010 | 11198 | -0.008 | -0.3609 | No | ||
| 78 | MMP16 | 2680139 | 11237 | -0.009 | -0.3629 | No | ||
| 79 | CNTN4 | 1780300 5570577 6370019 | 11264 | -0.009 | -0.3642 | No | ||
| 80 | DR1 | 5860750 | 11705 | -0.017 | -0.3879 | No | ||
| 81 | GABRA1 | 130372 | 11912 | -0.021 | -0.3989 | No | ||
| 82 | ZIC2 | 1410408 | 11949 | -0.021 | -0.4006 | No | ||
| 83 | CRK | 1230162 4780128 | 11967 | -0.022 | -0.4013 | No | ||
| 84 | MITF | 380056 | 12042 | -0.024 | -0.4051 | No | ||
| 85 | INSM1 | 3450671 | 12097 | -0.025 | -0.4078 | No | ||
| 86 | HS6ST2 | 520133 5550603 | 12302 | -0.029 | -0.4186 | No | ||
| 87 | SLC25A37 | 6520341 460471 | 12376 | -0.031 | -0.4222 | No | ||
| 88 | THRB | 1780673 | 12379 | -0.031 | -0.4220 | No | ||
| 89 | KCNJ3 | 50184 | 12469 | -0.033 | -0.4265 | No | ||
| 90 | GOLGA4 | 460300 | 12780 | -0.042 | -0.4429 | No | ||
| 91 | E2F7 | 4200056 | 12939 | -0.046 | -0.4510 | No | ||
| 92 | NR2F2 | 3170609 3310577 | 12988 | -0.048 | -0.4532 | No | ||
| 93 | NEGR1 | 1940731 5130170 | 13122 | -0.053 | -0.4599 | No | ||
| 94 | TCEA1 | 5340070 | 13167 | -0.054 | -0.4617 | No | ||
| 95 | DVL2 | 6110162 | 13171 | -0.055 | -0.4614 | No | ||
| 96 | SLITRK2 | 2640020 | 13173 | -0.055 | -0.4609 | No | ||
| 97 | FRAS1 | 5080170 5670347 | 13275 | -0.059 | -0.4658 | No | ||
| 98 | HIPK1 | 110193 | 13682 | -0.076 | -0.4871 | No | ||
| 99 | E2F3 | 50162 460180 | 13868 | -0.086 | -0.4963 | No | ||
| 100 | HMGB2 | 2640603 | 13950 | -0.091 | -0.4998 | No | ||
| 101 | LRRC4 | 3850041 | 13961 | -0.092 | -0.4994 | No | ||
| 102 | SORCS3 | 5700309 | 13992 | -0.094 | -0.5001 | No | ||
| 103 | PCTK2 | 5700673 | 14026 | -0.096 | -0.5010 | No | ||
| 104 | AKAP2 | 5550347 | 14074 | -0.099 | -0.5026 | No | ||
| 105 | SORCS1 | 60411 5890373 | 14120 | -0.102 | -0.5040 | No | ||
| 106 | HNRPH2 | 2630040 5720671 | 14185 | -0.107 | -0.5065 | No | ||
| 107 | RARG | 6760136 | 14266 | -0.114 | -0.5097 | No | ||
| 108 | HIP2 | 2810095 3990369 4120301 | 14405 | -0.126 | -0.5160 | No | ||
| 109 | NR4A3 | 2900021 5860095 5910039 | 14540 | -0.138 | -0.5219 | No | ||
| 110 | QKI | 5220093 6130044 | 14544 | -0.139 | -0.5207 | No | ||
| 111 | ATP11A | 1240717 5570603 | 14568 | -0.141 | -0.5206 | No | ||
| 112 | SLITRK3 | 5910041 | 14698 | -0.157 | -0.5260 | No | ||
| 113 | LRP4 | 1740253 | 14885 | -0.184 | -0.5343 | No | ||
| 114 | NFKBIZ | 2970450 | 14989 | -0.200 | -0.5380 | No | ||
| 115 | NLK | 2030010 2450041 | 15161 | -0.233 | -0.5450 | No | ||
| 116 | STC1 | 360161 | 15255 | -0.253 | -0.5476 | No | ||
| 117 | CASK | 2340215 3290576 6770215 | 15478 | -0.302 | -0.5567 | No | ||
| 118 | RBBP5 | 2760735 6760164 | 15715 | -0.374 | -0.5658 | No | ||
| 119 | DNAJC6 | 4590438 5900070 | 15733 | -0.378 | -0.5631 | No | ||
| 120 | KCNN2 | 990025 3520524 | 15883 | -0.431 | -0.5670 | Yes | ||
| 121 | TIMP3 | 1450504 1980270 | 15927 | -0.443 | -0.5650 | Yes | ||
| 122 | PSCD3 | 730301 | 15998 | -0.466 | -0.5643 | Yes | ||
| 123 | NR3C1 | 630563 | 16015 | -0.470 | -0.5606 | Yes | ||
| 124 | STRN3 | 1450093 6200685 | 16018 | -0.471 | -0.5561 | Yes | ||
| 125 | STAT1 | 6510204 6590553 | 16041 | -0.480 | -0.5527 | Yes | ||
| 126 | CIT | 2370601 | 16181 | -0.528 | -0.5551 | Yes | ||
| 127 | PHF16 | 50373 7200161 | 16190 | -0.533 | -0.5503 | Yes | ||
| 128 | PPP3R1 | 1190041 1570487 | 16230 | -0.549 | -0.5471 | Yes | ||
| 129 | CUGBP2 | 6180121 6840139 | 16339 | -0.592 | -0.5472 | Yes | ||
| 130 | ERBB2IP | 580253 1090672 | 16411 | -0.621 | -0.5450 | Yes | ||
| 131 | TNFRSF19L | 1170168 1190358 5670270 | 16476 | -0.650 | -0.5422 | Yes | ||
| 132 | FBXO33 | 1510504 | 16499 | -0.664 | -0.5369 | Yes | ||
| 133 | CYGB | 870347 | 16519 | -0.672 | -0.5314 | Yes | ||
| 134 | ITGB1 | 5080156 6270528 | 16739 | -0.801 | -0.5355 | Yes | ||
| 135 | YAF2 | 940053 | 16761 | -0.810 | -0.5288 | Yes | ||
| 136 | PCAF | 2230161 2570369 6550451 | 16765 | -0.812 | -0.5211 | Yes | ||
| 137 | YWHAZ | 1230717 | 16788 | -0.828 | -0.5142 | Yes | ||
| 138 | KRIT1 | 2640500 4230435 4730184 | 16823 | -0.851 | -0.5078 | Yes | ||
| 139 | TRIM33 | 580619 2230280 3990433 6200747 | 16870 | -0.879 | -0.5018 | Yes | ||
| 140 | PDK1 | 5290427 | 16984 | -0.956 | -0.4986 | Yes | ||
| 141 | ACTL6A | 5220543 6290541 2450093 4590332 5270722 | 17037 | -0.982 | -0.4919 | Yes | ||
| 142 | ARNTL | 3170463 | 17281 | -1.129 | -0.4941 | Yes | ||
| 143 | KCTD9 | 780577 | 17341 | -1.180 | -0.4859 | Yes | ||
| 144 | PHF2 | 4060300 | 17353 | -1.188 | -0.4749 | Yes | ||
| 145 | MIB1 | 3800537 | 17399 | -1.228 | -0.4654 | Yes | ||
| 146 | SOX4 | 2260091 | 17476 | -1.291 | -0.4570 | Yes | ||
| 147 | EIF4ENIF1 | 2060341 | 17486 | -1.305 | -0.4448 | Yes | ||
| 148 | FYCO1 | 3990112 | 17534 | -1.351 | -0.4343 | Yes | ||
| 149 | GSPT1 | 5420050 | 17593 | -1.408 | -0.4237 | Yes | ||
| 150 | SAR1B | 2510273 | 17625 | -1.442 | -0.4114 | Yes | ||
| 151 | DDX3X | 2190020 | 17667 | -1.484 | -0.3992 | Yes | ||
| 152 | ZHX1 | 2100110 2120286 | 17766 | -1.596 | -0.3890 | Yes | ||
| 153 | MECP2 | 1940450 | 17781 | -1.615 | -0.3741 | Yes | ||
| 154 | ELOVL6 | 5340746 | 17800 | -1.633 | -0.3593 | Yes | ||
| 155 | BCL11B | 2680673 | 17870 | -1.704 | -0.3464 | Yes | ||
| 156 | SENP1 | 6100537 | 17960 | -1.792 | -0.3339 | Yes | ||
| 157 | TSGA14 | 6660465 | 17987 | -1.829 | -0.3175 | Yes | ||
| 158 | KRAS | 2060170 | 17995 | -1.838 | -0.3001 | Yes | ||
| 159 | RAP2C | 1690132 | 18028 | -1.868 | -0.2837 | Yes | ||
| 160 | ANKRD10 | 1240025 | 18068 | -1.906 | -0.2673 | Yes | ||
| 161 | UBE2D3 | 3190452 | 18074 | -1.915 | -0.2489 | Yes | ||
| 162 | SFMBT1 | 2360450 2370070 | 18079 | -1.923 | -0.2305 | Yes | ||
| 163 | MAP3K7 | 6040068 | 18082 | -1.929 | -0.2119 | Yes | ||
| 164 | ETF1 | 6770075 | 18136 | -2.016 | -0.1952 | Yes | ||
| 165 | VAMP4 | 4150102 | 18168 | -2.064 | -0.1768 | Yes | ||
| 166 | STK4 | 2640152 | 18189 | -2.091 | -0.1576 | Yes | ||
| 167 | RAB14 | 6860139 | 18296 | -2.272 | -0.1413 | Yes | ||
| 168 | PSMD7 | 2030619 6220594 | 18323 | -2.337 | -0.1200 | Yes | ||
| 169 | APRIN | 2810050 | 18392 | -2.494 | -0.0995 | Yes | ||
| 170 | MTMR12 | 7000181 | 18485 | -2.755 | -0.0777 | Yes | ||
| 171 | KPNB1 | 1690138 | 18506 | -2.828 | -0.0513 | Yes | ||
| 172 | NDEL1 | 2370465 | 18522 | -2.900 | -0.0240 | Yes | ||
| 173 | SKP1A | 2450102 | 18536 | -2.990 | 0.0043 | Yes |

