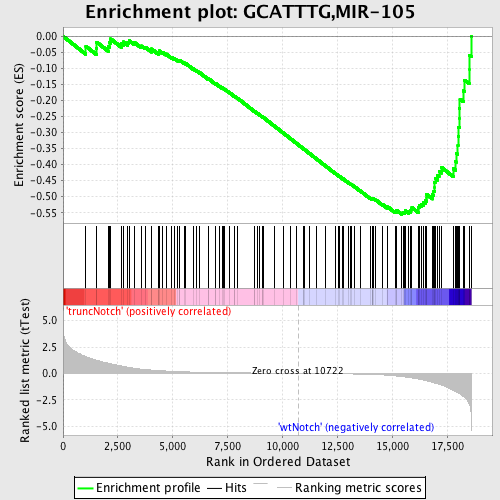
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | GCATTTG,MIR-105 |
| Enrichment Score (ES) | -0.55543786 |
| Normalized Enrichment Score (NES) | -1.6845056 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.016120398 |
| FWER p-Value | 0.143 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SNIP1 | 5360092 | 1039 | 1.585 | -0.0312 | No | ||
| 2 | OAZ2 | 2810110 | 1521 | 1.257 | -0.0374 | No | ||
| 3 | HDHD2 | 2850411 6940168 | 1533 | 1.249 | -0.0183 | No | ||
| 4 | TNFRSF12A | 4810253 | 2071 | 0.963 | -0.0322 | No | ||
| 5 | SON | 3120148 5860239 7040403 | 2102 | 0.945 | -0.0189 | No | ||
| 6 | COPS8 | 2810541 | 2172 | 0.912 | -0.0083 | No | ||
| 7 | CAMK2G | 630097 | 2653 | 0.695 | -0.0233 | No | ||
| 8 | ITM2B | 1850482 5860671 6200014 | 2739 | 0.656 | -0.0175 | No | ||
| 9 | SGCD | 2940685 5080753 | 2946 | 0.584 | -0.0195 | No | ||
| 10 | PTPN13 | 2060681 | 3003 | 0.565 | -0.0136 | No | ||
| 11 | KPNA4 | 510369 | 3238 | 0.482 | -0.0186 | No | ||
| 12 | EIF3S10 | 1190079 3840176 | 3569 | 0.403 | -0.0301 | No | ||
| 13 | MXI1 | 5050064 5130484 | 3745 | 0.363 | -0.0339 | No | ||
| 14 | IGF1 | 1990193 3130377 3290280 | 4031 | 0.315 | -0.0443 | No | ||
| 15 | FZD7 | 7050706 | 4033 | 0.315 | -0.0394 | No | ||
| 16 | RB1CC1 | 510494 7100072 | 4347 | 0.266 | -0.0522 | No | ||
| 17 | SP8 | 4060576 | 4366 | 0.264 | -0.0490 | No | ||
| 18 | FLRT2 | 4200035 | 4379 | 0.262 | -0.0455 | No | ||
| 19 | GAD1 | 2360035 3140167 | 4529 | 0.240 | -0.0498 | No | ||
| 20 | HTR2A | 360037 6330255 | 4694 | 0.222 | -0.0551 | No | ||
| 21 | PRKD1 | 1410706 5090438 | 4956 | 0.195 | -0.0662 | No | ||
| 22 | SOCS5 | 3830398 7100093 | 5084 | 0.184 | -0.0701 | No | ||
| 23 | HTR2C | 380497 | 5216 | 0.172 | -0.0745 | No | ||
| 24 | CUGBP1 | 450292 510022 7050176 7050215 | 5294 | 0.166 | -0.0761 | No | ||
| 25 | CDH2 | 520435 2450451 2760025 6650477 | 5313 | 0.164 | -0.0744 | No | ||
| 26 | BMP1 | 380594 2940576 3710593 | 5516 | 0.149 | -0.0830 | No | ||
| 27 | CREB5 | 2320368 | 5586 | 0.144 | -0.0845 | No | ||
| 28 | MYLIP | 50717 4050672 | 5932 | 0.123 | -0.1012 | No | ||
| 29 | KLF12 | 1660095 4810288 5340546 6520286 | 6072 | 0.115 | -0.1069 | No | ||
| 30 | POU4F1 | 2260315 | 6227 | 0.108 | -0.1135 | No | ||
| 31 | MAP3K2 | 380270 | 6608 | 0.091 | -0.1326 | No | ||
| 32 | PTPRG | 2650524 1340022 | 6633 | 0.090 | -0.1325 | No | ||
| 33 | ETV1 | 70735 2940603 5080463 | 6938 | 0.079 | -0.1477 | No | ||
| 34 | SRGAP1 | 3940253 | 6963 | 0.077 | -0.1478 | No | ||
| 35 | AMOTL2 | 4730082 | 7137 | 0.072 | -0.1560 | No | ||
| 36 | FAIM2 | 2100014 | 7242 | 0.069 | -0.1605 | No | ||
| 37 | MEF2C | 670025 780338 | 7314 | 0.067 | -0.1633 | No | ||
| 38 | SOX6 | 2940458 4070601 6840717 | 7362 | 0.066 | -0.1648 | No | ||
| 39 | CPXM2 | 4560452 | 7562 | 0.060 | -0.1746 | No | ||
| 40 | LRRC16 | 360022 6450070 | 7800 | 0.054 | -0.1866 | No | ||
| 41 | ARHGEF7 | 1690441 | 7815 | 0.054 | -0.1865 | No | ||
| 42 | PRDM8 | 840746 | 7950 | 0.050 | -0.1929 | No | ||
| 43 | GABRB3 | 4150164 | 8713 | 0.034 | -0.2336 | No | ||
| 44 | BTBD11 | 6650154 | 8730 | 0.034 | -0.2339 | No | ||
| 45 | SSPN | 4670091 | 8878 | 0.031 | -0.2414 | No | ||
| 46 | PRPF4B | 940181 2570300 | 8962 | 0.029 | -0.2454 | No | ||
| 47 | ZFR | 5910647 | 9092 | 0.027 | -0.2520 | No | ||
| 48 | TIPARP | 50397 | 9093 | 0.027 | -0.2515 | No | ||
| 49 | ACVR2A | 6110647 | 9148 | 0.026 | -0.2540 | No | ||
| 50 | CCDC3 | 7000441 | 9647 | 0.018 | -0.2807 | No | ||
| 51 | SP4 | 3850176 | 10063 | 0.011 | -0.3029 | No | ||
| 52 | ABCC12 | 5550026 | 10363 | 0.006 | -0.3190 | No | ||
| 53 | THOC1 | 110095 | 10641 | 0.001 | -0.3340 | No | ||
| 54 | SLITRK5 | 6590292 | 10963 | -0.004 | -0.3513 | No | ||
| 55 | SP3 | 3840338 | 10973 | -0.004 | -0.3517 | No | ||
| 56 | PRICKLE2 | 780408 | 11010 | -0.005 | -0.3536 | No | ||
| 57 | L3MBTL3 | 3800113 6660068 | 11214 | -0.008 | -0.3644 | No | ||
| 58 | ZIC3 | 380020 | 11532 | -0.013 | -0.3813 | No | ||
| 59 | CRK | 1230162 4780128 | 11967 | -0.022 | -0.4044 | No | ||
| 60 | EPHA4 | 460750 | 12424 | -0.032 | -0.4286 | No | ||
| 61 | SATB2 | 2470181 | 12557 | -0.035 | -0.4352 | No | ||
| 62 | NF2 | 4150735 6450139 | 12588 | -0.036 | -0.4362 | No | ||
| 63 | RAD52 | 110093 | 12754 | -0.041 | -0.4445 | No | ||
| 64 | SOCS7 | 1400372 6860671 | 12756 | -0.041 | -0.4439 | No | ||
| 65 | PKD1L2 | 1780341 5420609 | 13005 | -0.048 | -0.4566 | No | ||
| 66 | PFTK1 | 1780181 | 13110 | -0.052 | -0.4614 | No | ||
| 67 | AMOT | 1940121 2940647 6590093 | 13140 | -0.053 | -0.4621 | No | ||
| 68 | FRAS1 | 5080170 5670347 | 13275 | -0.059 | -0.4684 | No | ||
| 69 | GLDC | 1170324 | 13543 | -0.070 | -0.4818 | No | ||
| 70 | CRKRS | 510537 2510551 | 13994 | -0.094 | -0.5046 | No | ||
| 71 | ITGA6 | 3830129 | 14095 | -0.100 | -0.5084 | No | ||
| 72 | TJP1 | 6350184 | 14107 | -0.101 | -0.5074 | No | ||
| 73 | NRXN1 | 460408 5340270 6370066 | 14110 | -0.101 | -0.5059 | No | ||
| 74 | USP15 | 610592 3520504 | 14148 | -0.104 | -0.5063 | No | ||
| 75 | ST8SIA3 | 6940427 | 14259 | -0.114 | -0.5105 | No | ||
| 76 | CDKN1B | 3800025 6450044 | 14572 | -0.142 | -0.5251 | No | ||
| 77 | EIF2C1 | 6900551 | 14773 | -0.168 | -0.5333 | No | ||
| 78 | OLIG3 | 1050603 | 14794 | -0.171 | -0.5316 | No | ||
| 79 | ARRDC3 | 4610390 | 15141 | -0.230 | -0.5467 | No | ||
| 80 | FNDC3A | 2690097 3290044 | 15178 | -0.236 | -0.5450 | No | ||
| 81 | PTTG1IP | 4670441 | 15185 | -0.238 | -0.5415 | No | ||
| 82 | PRDM2 | 780278 1940692 | 15443 | -0.292 | -0.5508 | Yes | ||
| 83 | PHF12 | 870400 3130687 | 15491 | -0.306 | -0.5486 | Yes | ||
| 84 | GPR85 | 5130750 5910020 | 15572 | -0.327 | -0.5477 | Yes | ||
| 85 | RAB22A | 110500 3830707 | 15584 | -0.331 | -0.5431 | Yes | ||
| 86 | SFRS10 | 6130528 6400438 7040112 | 15755 | -0.384 | -0.5463 | Yes | ||
| 87 | FOXK1 | 2640373 3290136 | 15834 | -0.411 | -0.5440 | Yes | ||
| 88 | YTHDF3 | 4540632 4670717 7040609 | 15856 | -0.421 | -0.5385 | Yes | ||
| 89 | GABARAPL2 | 610204 3360047 | 15898 | -0.434 | -0.5339 | Yes | ||
| 90 | CPEB1 | 3190524 | 16189 | -0.533 | -0.5412 | Yes | ||
| 91 | PHF16 | 50373 7200161 | 16190 | -0.533 | -0.5328 | Yes | ||
| 92 | PPP3R1 | 1190041 1570487 | 16230 | -0.549 | -0.5262 | Yes | ||
| 93 | CUGBP2 | 6180121 6840139 | 16339 | -0.592 | -0.5228 | Yes | ||
| 94 | DICER1 | 540286 | 16417 | -0.623 | -0.5171 | Yes | ||
| 95 | PDE1B | 3440066 | 16506 | -0.667 | -0.5114 | Yes | ||
| 96 | GOSR1 | 730279 | 16565 | -0.690 | -0.5036 | Yes | ||
| 97 | EXTL2 | 5050433 | 16568 | -0.690 | -0.4929 | Yes | ||
| 98 | LDB1 | 5270601 | 16834 | -0.855 | -0.4937 | Yes | ||
| 99 | RYBP | 60746 | 16895 | -0.894 | -0.4829 | Yes | ||
| 100 | EIF4G2 | 3800575 6860184 | 16927 | -0.918 | -0.4701 | Yes | ||
| 101 | GRPEL2 | 6900154 | 16932 | -0.919 | -0.4558 | Yes | ||
| 102 | PCCA | 3390400 | 16958 | -0.935 | -0.4425 | Yes | ||
| 103 | TPM3 | 670600 3170296 5670167 6650471 | 17068 | -0.998 | -0.4327 | Yes | ||
| 104 | TM9SF2 | 430332 6900156 | 17137 | -1.023 | -0.4202 | Yes | ||
| 105 | TEX2 | 1740504 2480102 3060494 | 17252 | -1.102 | -0.4090 | Yes | ||
| 106 | MECP2 | 1940450 | 17781 | -1.615 | -0.4121 | Yes | ||
| 107 | MORF4L2 | 6450133 | 17889 | -1.716 | -0.3909 | Yes | ||
| 108 | H3F3A | 2900086 | 17914 | -1.745 | -0.3647 | Yes | ||
| 109 | KRAS | 2060170 | 17995 | -1.838 | -0.3401 | Yes | ||
| 110 | AMMECR1 | 3800435 | 18003 | -1.846 | -0.3114 | Yes | ||
| 111 | RAP2C | 1690132 | 18028 | -1.868 | -0.2833 | Yes | ||
| 112 | TULP4 | 2320364 | 18067 | -1.905 | -0.2553 | Yes | ||
| 113 | NCOA1 | 3610438 | 18073 | -1.911 | -0.2255 | Yes | ||
| 114 | HNRPK | 1190348 2060411 2570092 6840551 | 18088 | -1.938 | -0.1957 | Yes | ||
| 115 | COPS4 | 4230497 6940324 | 18254 | -2.195 | -0.1701 | Yes | ||
| 116 | H3F3B | 1410300 | 18304 | -2.281 | -0.1368 | Yes | ||
| 117 | OGT | 2360131 4610333 | 18507 | -2.829 | -0.1031 | Yes | ||
| 118 | NEDD4L | 6380368 | 18511 | -2.841 | -0.0586 | Yes | ||
| 119 | SLC31A1 | 4590161 6940154 | 18606 | -4.075 | 0.0005 | Yes |

