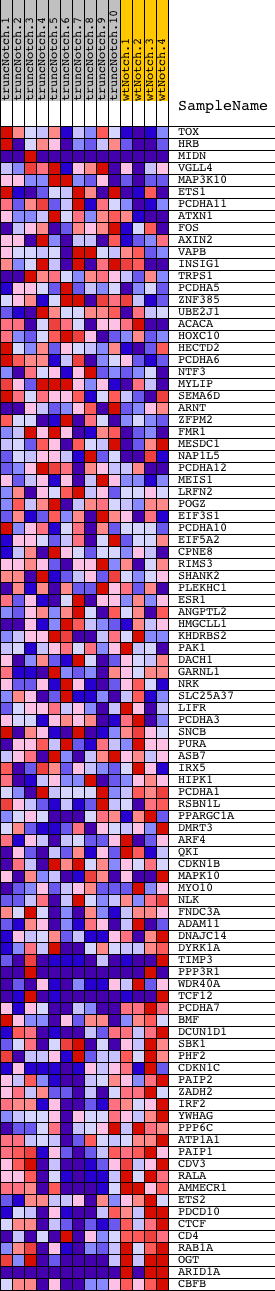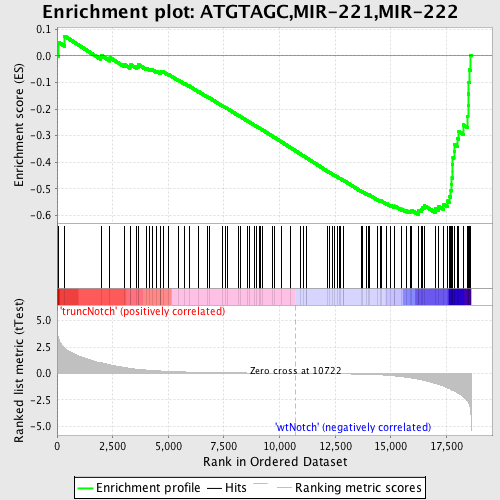
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | ATGTAGC,MIR-221,MIR-222 |
| Enrichment Score (ES) | -0.59636205 |
| Normalized Enrichment Score (NES) | -1.7402568 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.009136908 |
| FWER p-Value | 0.066 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TOX | 3440053 | 50 | 3.421 | 0.0520 | No | ||
| 2 | HRB | 1660397 6110398 | 341 | 2.382 | 0.0745 | No | ||
| 3 | MIDN | 2260132 5220377 | 1972 | 1.000 | 0.0025 | No | ||
| 4 | VGLL4 | 6860463 | 2375 | 0.818 | -0.0062 | No | ||
| 5 | MAP3K10 | 3360168 4850180 | 3023 | 0.555 | -0.0322 | No | ||
| 6 | ETS1 | 5270278 6450717 6620465 | 3277 | 0.470 | -0.0384 | No | ||
| 7 | PCDHA11 | 1510253 | 3317 | 0.459 | -0.0331 | No | ||
| 8 | ATXN1 | 5550156 | 3574 | 0.402 | -0.0405 | No | ||
| 9 | FOS | 1850315 | 3645 | 0.386 | -0.0381 | No | ||
| 10 | AXIN2 | 3850131 | 3666 | 0.383 | -0.0331 | No | ||
| 11 | VAPB | 1660451 6130333 | 4029 | 0.315 | -0.0476 | No | ||
| 12 | INSIG1 | 1500332 3450458 4540301 | 4153 | 0.292 | -0.0495 | No | ||
| 13 | TRPS1 | 870100 | 4270 | 0.274 | -0.0514 | No | ||
| 14 | PCDHA5 | 2970347 | 4473 | 0.248 | -0.0584 | No | ||
| 15 | ZNF385 | 450176 4810358 | 4632 | 0.230 | -0.0632 | No | ||
| 16 | UBE2J1 | 4070133 | 4660 | 0.227 | -0.0610 | No | ||
| 17 | ACACA | 2490612 2680369 | 4667 | 0.226 | -0.0578 | No | ||
| 18 | HOXC10 | 5360170 | 4767 | 0.215 | -0.0597 | No | ||
| 19 | HECTD2 | 2680014 3060689 | 4997 | 0.192 | -0.0690 | No | ||
| 20 | PCDHA6 | 780333 2370280 | 5460 | 0.153 | -0.0914 | No | ||
| 21 | NTF3 | 5890131 | 5743 | 0.134 | -0.1045 | No | ||
| 22 | MYLIP | 50717 4050672 | 5932 | 0.123 | -0.1127 | No | ||
| 23 | SEMA6D | 4050324 5860138 6350307 | 6367 | 0.101 | -0.1345 | No | ||
| 24 | ARNT | 1170672 5670711 | 6740 | 0.086 | -0.1533 | No | ||
| 25 | ZFPM2 | 460072 | 6833 | 0.082 | -0.1569 | No | ||
| 26 | FMR1 | 5050075 | 7450 | 0.063 | -0.1892 | No | ||
| 27 | MESDC1 | 2030019 | 7551 | 0.061 | -0.1936 | No | ||
| 28 | NAP1L5 | 360576 | 7661 | 0.057 | -0.1986 | No | ||
| 29 | PCDHA12 | 2810215 | 8150 | 0.046 | -0.2242 | No | ||
| 30 | MEIS1 | 1400575 | 8223 | 0.044 | -0.2274 | No | ||
| 31 | LRFN2 | 6450411 | 8568 | 0.037 | -0.2453 | No | ||
| 32 | POGZ | 870348 7040286 | 8639 | 0.036 | -0.2485 | No | ||
| 33 | EIF3S1 | 6130368 6770044 | 8880 | 0.031 | -0.2610 | No | ||
| 34 | PCDHA10 | 3830725 | 8960 | 0.029 | -0.2648 | No | ||
| 35 | EIF5A2 | 7040139 | 9111 | 0.027 | -0.2725 | No | ||
| 36 | CPNE8 | 7040039 | 9151 | 0.026 | -0.2742 | No | ||
| 37 | RIMS3 | 3890563 | 9228 | 0.025 | -0.2779 | No | ||
| 38 | SHANK2 | 1500452 | 9665 | 0.017 | -0.3011 | No | ||
| 39 | PLEKHC1 | 2650066 | 9777 | 0.015 | -0.3069 | No | ||
| 40 | ESR1 | 4060372 5860193 | 10104 | 0.010 | -0.3243 | No | ||
| 41 | ANGPTL2 | 6980152 6760162 | 10493 | 0.004 | -0.3452 | No | ||
| 42 | HMGCLL1 | 2570091 | 10941 | -0.004 | -0.3693 | No | ||
| 43 | KHDRBS2 | 3520717 | 11079 | -0.006 | -0.3766 | No | ||
| 44 | PAK1 | 4540315 | 11187 | -0.008 | -0.3822 | No | ||
| 45 | DACH1 | 2450593 5700592 | 11221 | -0.008 | -0.3839 | No | ||
| 46 | GARNL1 | 5670398 | 12165 | -0.026 | -0.4344 | No | ||
| 47 | NRK | 4590609 | 12252 | -0.028 | -0.4386 | No | ||
| 48 | SLC25A37 | 6520341 460471 | 12376 | -0.031 | -0.4447 | No | ||
| 49 | LIFR | 3360068 3830102 | 12452 | -0.033 | -0.4483 | No | ||
| 50 | PCDHA3 | 3840402 | 12586 | -0.036 | -0.4549 | No | ||
| 51 | SNCB | 3360133 | 12691 | -0.039 | -0.4599 | No | ||
| 52 | PURA | 3870156 | 12716 | -0.040 | -0.4605 | No | ||
| 53 | ASB7 | 520242 2450088 3990563 4280538 | 12882 | -0.044 | -0.4687 | No | ||
| 54 | IRX5 | 2630494 | 12888 | -0.044 | -0.4683 | No | ||
| 55 | HIPK1 | 110193 | 13682 | -0.076 | -0.5099 | No | ||
| 56 | PCDHA1 | 940026 540484 | 13707 | -0.077 | -0.5099 | No | ||
| 57 | RSBN1L | 50647 | 13903 | -0.088 | -0.5191 | No | ||
| 58 | PPARGC1A | 4670040 | 13995 | -0.094 | -0.5225 | No | ||
| 59 | DMRT3 | 840338 | 14022 | -0.096 | -0.5224 | No | ||
| 60 | ARF4 | 3190661 | 14414 | -0.127 | -0.5414 | No | ||
| 61 | QKI | 5220093 6130044 | 14544 | -0.139 | -0.5462 | No | ||
| 62 | CDKN1B | 3800025 6450044 | 14572 | -0.142 | -0.5454 | No | ||
| 63 | MAPK10 | 6110193 | 14822 | -0.175 | -0.5560 | No | ||
| 64 | MYO10 | 3830576 5570092 | 14992 | -0.201 | -0.5619 | No | ||
| 65 | NLK | 2030010 2450041 | 15161 | -0.233 | -0.5673 | No | ||
| 66 | FNDC3A | 2690097 3290044 | 15178 | -0.236 | -0.5644 | No | ||
| 67 | ADAM11 | 1050008 3130494 | 15477 | -0.302 | -0.5756 | No | ||
| 68 | DNAJC14 | 6420369 | 15690 | -0.366 | -0.5812 | No | ||
| 69 | DYRK1A | 3190181 | 15866 | -0.423 | -0.5839 | No | ||
| 70 | TIMP3 | 1450504 1980270 | 15927 | -0.443 | -0.5801 | No | ||
| 71 | PPP3R1 | 1190041 1570487 | 16230 | -0.549 | -0.5876 | Yes | ||
| 72 | WDR40A | 2190167 | 16254 | -0.557 | -0.5799 | Yes | ||
| 73 | TCF12 | 3610324 7000156 | 16383 | -0.608 | -0.5771 | Yes | ||
| 74 | PCDHA7 | 1850019 | 16415 | -0.622 | -0.5688 | Yes | ||
| 75 | BMF | 610102 | 16518 | -0.672 | -0.5636 | Yes | ||
| 76 | DCUN1D1 | 2360332 | 17007 | -0.972 | -0.5744 | Yes | ||
| 77 | SBK1 | 940452 | 17157 | -1.037 | -0.5658 | Yes | ||
| 78 | PHF2 | 4060300 | 17353 | -1.188 | -0.5573 | Yes | ||
| 79 | CDKN1C | 6520577 | 17535 | -1.353 | -0.5455 | Yes | ||
| 80 | PAIP2 | 4210441 | 17643 | -1.457 | -0.5279 | Yes | ||
| 81 | ZADH2 | 520438 | 17701 | -1.523 | -0.5067 | Yes | ||
| 82 | IRF2 | 5390044 | 17728 | -1.557 | -0.4832 | Yes | ||
| 83 | YWHAG | 3780341 | 17748 | -1.581 | -0.4589 | Yes | ||
| 84 | PPP6C | 2450670 | 17776 | -1.613 | -0.4345 | Yes | ||
| 85 | ATP1A1 | 5670451 | 17777 | -1.613 | -0.4087 | Yes | ||
| 86 | PAIP1 | 3870576 | 17791 | -1.625 | -0.3835 | Yes | ||
| 87 | CDV3 | 1940577 | 17840 | -1.671 | -0.3593 | Yes | ||
| 88 | RALA | 2680471 | 17860 | -1.692 | -0.3333 | Yes | ||
| 89 | AMMECR1 | 3800435 | 18003 | -1.846 | -0.3114 | Yes | ||
| 90 | ETS2 | 360451 | 18058 | -1.897 | -0.2840 | Yes | ||
| 91 | PDCD10 | 4760500 | 18256 | -2.201 | -0.2594 | Yes | ||
| 92 | CTCF | 5340017 | 18444 | -2.618 | -0.2276 | Yes | ||
| 93 | CD4 | 1090010 | 18469 | -2.701 | -0.1857 | Yes | ||
| 94 | RAB1A | 2370671 | 18476 | -2.727 | -0.1424 | Yes | ||
| 95 | OGT | 2360131 4610333 | 18507 | -2.829 | -0.0988 | Yes | ||
| 96 | ARID1A | 2630022 1690551 4810110 | 18538 | -2.995 | -0.0525 | Yes | ||
| 97 | CBFB | 1850551 2650035 | 18587 | -3.540 | 0.0016 | Yes |