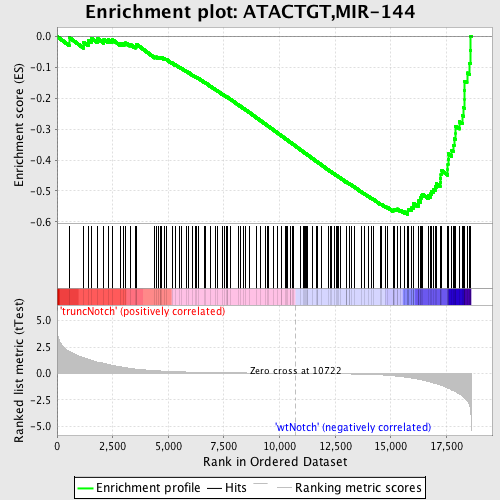
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | ATACTGT,MIR-144 |
| Enrichment Score (ES) | -0.5763618 |
| Normalized Enrichment Score (NES) | -1.7778151 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.006463589 |
| FWER p-Value | 0.032 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | AGRN | 6770152 | 542 | 2.084 | -0.0030 | No | ||
| 2 | ARID2 | 110647 5390435 | 1184 | 1.479 | -0.0190 | No | ||
| 3 | ATP5G2 | 840068 | 1398 | 1.340 | -0.0136 | No | ||
| 4 | HDHD2 | 2850411 6940168 | 1533 | 1.249 | -0.0050 | No | ||
| 5 | MYST3 | 5270500 | 1802 | 1.077 | -0.0059 | No | ||
| 6 | SON | 3120148 5860239 7040403 | 2102 | 0.945 | -0.0101 | No | ||
| 7 | PIM1 | 630047 | 2298 | 0.856 | -0.0099 | No | ||
| 8 | CPEB2 | 4760338 | 2471 | 0.773 | -0.0094 | No | ||
| 9 | DLG5 | 450215 | 2839 | 0.620 | -0.0214 | No | ||
| 10 | STARD8 | 1780047 | 2979 | 0.571 | -0.0217 | No | ||
| 11 | LHX2 | 3610463 | 3089 | 0.532 | -0.0209 | No | ||
| 12 | ETS1 | 5270278 6450717 6620465 | 3277 | 0.470 | -0.0251 | No | ||
| 13 | NIN | 3450576 5080048 | 3507 | 0.414 | -0.0322 | No | ||
| 14 | PDE7B | 5700056 | 3547 | 0.408 | -0.0292 | No | ||
| 15 | ATXN1 | 5550156 | 3574 | 0.402 | -0.0255 | No | ||
| 16 | PTHLH | 5290739 | 4393 | 0.260 | -0.0665 | No | ||
| 17 | HOXA10 | 6110397 7100458 | 4449 | 0.252 | -0.0663 | No | ||
| 18 | PTGFRN | 4120524 | 4557 | 0.237 | -0.0691 | No | ||
| 19 | JPH1 | 610739 | 4629 | 0.230 | -0.0700 | No | ||
| 20 | EDAR | 6220670 | 4656 | 0.227 | -0.0686 | No | ||
| 21 | NFE2L2 | 2810619 3390162 | 4711 | 0.221 | -0.0687 | No | ||
| 22 | SLC12A2 | 4610019 6040672 | 4813 | 0.210 | -0.0715 | No | ||
| 23 | PAPPA | 4230463 | 4896 | 0.202 | -0.0734 | No | ||
| 24 | BACH2 | 6760131 | 5165 | 0.177 | -0.0857 | No | ||
| 25 | CDH2 | 520435 2450451 2760025 6650477 | 5313 | 0.164 | -0.0915 | No | ||
| 26 | ARID5B | 110403 | 5496 | 0.151 | -0.0995 | No | ||
| 27 | WIF1 | 7100184 | 5600 | 0.143 | -0.1033 | No | ||
| 28 | SMPD3 | 520164 | 5815 | 0.130 | -0.1132 | No | ||
| 29 | CALCRL | 4280035 | 5913 | 0.124 | -0.1169 | No | ||
| 30 | DGCR2 | 780041 4010731 | 6104 | 0.114 | -0.1257 | No | ||
| 31 | DMD | 1740041 3990332 | 6236 | 0.107 | -0.1315 | No | ||
| 32 | TRIB1 | 2320435 | 6277 | 0.106 | -0.1323 | No | ||
| 33 | SEMA6D | 4050324 5860138 6350307 | 6367 | 0.101 | -0.1359 | No | ||
| 34 | STAG1 | 1190300 1400722 4590100 | 6607 | 0.091 | -0.1477 | No | ||
| 35 | ST6GALNAC3 | 4070563 | 6663 | 0.089 | -0.1495 | No | ||
| 36 | CBX1 | 5080408 1980239 | 6915 | 0.079 | -0.1621 | No | ||
| 37 | SFRP1 | 6980315 | 7111 | 0.073 | -0.1717 | No | ||
| 38 | PAFAH1B1 | 4230333 6420121 6450066 | 7219 | 0.070 | -0.1767 | No | ||
| 39 | FMR1 | 5050075 | 7450 | 0.063 | -0.1883 | No | ||
| 40 | SEMA6A | 2690138 3060021 | 7533 | 0.061 | -0.1920 | No | ||
| 41 | CDH11 | 1230133 2100180 4610110 | 7613 | 0.059 | -0.1955 | No | ||
| 42 | ANK2 | 6510546 | 7637 | 0.058 | -0.1960 | No | ||
| 43 | CPEB3 | 3940164 | 7648 | 0.058 | -0.1958 | No | ||
| 44 | ALDH1A3 | 2100270 | 7806 | 0.054 | -0.2037 | No | ||
| 45 | PNRC1 | 360025 | 8164 | 0.046 | -0.2224 | No | ||
| 46 | MEIS1 | 1400575 | 8223 | 0.044 | -0.2250 | No | ||
| 47 | ST18 | 5720537 | 8379 | 0.041 | -0.2329 | No | ||
| 48 | TTN | 2320161 4670056 6550026 | 8471 | 0.039 | -0.2373 | No | ||
| 49 | WDFY3 | 3610041 4560333 4570273 | 8643 | 0.036 | -0.2461 | No | ||
| 50 | LGR4 | 1660309 | 8977 | 0.029 | -0.2638 | No | ||
| 51 | RFX4 | 3710100 5340215 | 9162 | 0.026 | -0.2734 | No | ||
| 52 | TBX1 | 3710133 6590121 | 9348 | 0.023 | -0.2832 | No | ||
| 53 | SLITRK4 | 5910022 | 9451 | 0.021 | -0.2884 | No | ||
| 54 | MYO1E | 3140504 | 9494 | 0.020 | -0.2904 | No | ||
| 55 | FMN2 | 450463 | 9722 | 0.016 | -0.3025 | No | ||
| 56 | ESRRG | 4010181 | 9926 | 0.013 | -0.3134 | No | ||
| 57 | SP4 | 3850176 | 10063 | 0.011 | -0.3206 | No | ||
| 58 | UBE4A | 6100520 | 10072 | 0.011 | -0.3209 | No | ||
| 59 | ZCCHC2 | 460592 | 10248 | 0.008 | -0.3303 | No | ||
| 60 | RIN2 | 2450184 | 10293 | 0.007 | -0.3326 | No | ||
| 61 | MSX1 | 2650309 | 10361 | 0.006 | -0.3361 | No | ||
| 62 | PPARA | 2060026 | 10473 | 0.004 | -0.3421 | No | ||
| 63 | SMARCA1 | 4230315 | 10479 | 0.004 | -0.3423 | No | ||
| 64 | ELL2 | 6420091 | 10575 | 0.002 | -0.3474 | No | ||
| 65 | CAV2 | 5130286 5130563 | 10617 | 0.002 | -0.3496 | No | ||
| 66 | HNRPF | 670138 | 10939 | -0.004 | -0.3670 | No | ||
| 67 | GLCCI1 | 110091 2640082 2900692 3290070 6370014 | 11086 | -0.006 | -0.3748 | No | ||
| 68 | SS18 | 1940440 2900072 6900129 | 11119 | -0.007 | -0.3765 | No | ||
| 69 | FBXW11 | 6450632 | 11158 | -0.007 | -0.3784 | No | ||
| 70 | PCDH18 | 430010 | 11198 | -0.008 | -0.3804 | No | ||
| 71 | PAX3 | 50551 | 11247 | -0.009 | -0.3829 | No | ||
| 72 | RGS16 | 780091 | 11252 | -0.009 | -0.3830 | No | ||
| 73 | PANK1 | 670091 6220605 | 11470 | -0.012 | -0.3946 | No | ||
| 74 | MYT1 | 6770524 | 11674 | -0.016 | -0.4054 | No | ||
| 75 | FAM60A | 3940092 | 11704 | -0.017 | -0.4068 | No | ||
| 76 | PLAG1 | 1450142 3870139 6520039 | 11711 | -0.017 | -0.4069 | No | ||
| 77 | CXCL12 | 580546 4150750 4570068 | 11898 | -0.020 | -0.4167 | No | ||
| 78 | SNAP91 | 3170056 5690520 | 12210 | -0.027 | -0.4332 | No | ||
| 79 | NRP2 | 4070400 5860041 6650446 | 12309 | -0.030 | -0.4381 | No | ||
| 80 | AHCY | 4230605 | 12348 | -0.030 | -0.4398 | No | ||
| 81 | LIFR | 3360068 3830102 | 12452 | -0.033 | -0.4450 | No | ||
| 82 | IDH2 | 2680397 | 12572 | -0.035 | -0.4510 | No | ||
| 83 | RARB | 430139 1410138 | 12614 | -0.037 | -0.4527 | No | ||
| 84 | CBX4 | 70332 | 12642 | -0.038 | -0.4537 | No | ||
| 85 | PURA | 3870156 | 12716 | -0.040 | -0.4572 | No | ||
| 86 | NR2F2 | 3170609 3310577 | 12988 | -0.048 | -0.4712 | No | ||
| 87 | WTAP | 4010176 | 12992 | -0.048 | -0.4708 | No | ||
| 88 | FOSB | 1940142 | 13018 | -0.049 | -0.4715 | No | ||
| 89 | ATP2B2 | 3780397 | 13151 | -0.054 | -0.4780 | No | ||
| 90 | PTP4A1 | 130692 | 13161 | -0.054 | -0.4778 | No | ||
| 91 | CCNK | 6980048 | 13250 | -0.057 | -0.4818 | No | ||
| 92 | GDF10 | 4850082 | 13364 | -0.062 | -0.4872 | No | ||
| 93 | CAPZA1 | 3140390 | 13665 | -0.075 | -0.5025 | No | ||
| 94 | VKORC1L1 | 110220 1400050 | 13822 | -0.083 | -0.5099 | No | ||
| 95 | SORCS3 | 5700309 | 13992 | -0.094 | -0.5178 | No | ||
| 96 | RHOBTB1 | 1410010 | 14125 | -0.103 | -0.5237 | No | ||
| 97 | PALM2 | 4120068 7100398 | 14206 | -0.109 | -0.5266 | No | ||
| 98 | QKI | 5220093 6130044 | 14544 | -0.139 | -0.5431 | No | ||
| 99 | CCT2 | 1690113 | 14576 | -0.142 | -0.5430 | No | ||
| 100 | FBXO32 | 110037 610750 | 14782 | -0.170 | -0.5520 | No | ||
| 101 | RBM12 | 4570239 6040411 6660129 | 14852 | -0.179 | -0.5534 | No | ||
| 102 | PTPN9 | 3290408 | 15099 | -0.220 | -0.5640 | No | ||
| 103 | BRPF1 | 3170139 | 15112 | -0.224 | -0.5618 | No | ||
| 104 | ARRDC3 | 4610390 | 15141 | -0.230 | -0.5604 | No | ||
| 105 | FNDC3A | 2690097 3290044 | 15178 | -0.236 | -0.5594 | No | ||
| 106 | ZIC4 | 1500082 | 15277 | -0.256 | -0.5614 | No | ||
| 107 | ACSL4 | 510020 1240041 3190050 4540500 5570463 | 15284 | -0.258 | -0.5585 | No | ||
| 108 | CRSP2 | 1940059 | 15440 | -0.292 | -0.5632 | No | ||
| 109 | NEDD4 | 460736 1190280 | 15607 | -0.339 | -0.5679 | No | ||
| 110 | FBXL3 | 5910142 | 15764 | -0.387 | -0.5715 | Yes | ||
| 111 | RNF111 | 2970072 3140112 5130647 | 15769 | -0.389 | -0.5668 | Yes | ||
| 112 | CLK1 | 1780551 2100102 2340347 2480347 | 15772 | -0.390 | -0.5619 | Yes | ||
| 113 | SUCLA2 | 730632 1170292 | 15807 | -0.406 | -0.5586 | Yes | ||
| 114 | PDE4D | 2470528 6660014 | 15917 | -0.441 | -0.5589 | Yes | ||
| 115 | ALS2 | 3130546 6400070 | 15945 | -0.448 | -0.5547 | Yes | ||
| 116 | NARG1 | 5910563 6350095 | 16009 | -0.469 | -0.5522 | Yes | ||
| 117 | NR3C1 | 630563 | 16015 | -0.470 | -0.5465 | Yes | ||
| 118 | SLC7A11 | 2850138 | 16021 | -0.473 | -0.5408 | Yes | ||
| 119 | PPP3R1 | 1190041 1570487 | 16230 | -0.549 | -0.5451 | Yes | ||
| 120 | FOXO1A | 3120670 | 16243 | -0.553 | -0.5388 | Yes | ||
| 121 | HSF2 | 1400465 | 16257 | -0.559 | -0.5324 | Yes | ||
| 122 | CUGBP2 | 6180121 6840139 | 16339 | -0.592 | -0.5293 | Yes | ||
| 123 | ARFGEF1 | 6760494 | 16345 | -0.593 | -0.5221 | Yes | ||
| 124 | FUSIP1 | 520082 5390114 | 16369 | -0.602 | -0.5157 | Yes | ||
| 125 | PUM1 | 6130500 | 16425 | -0.627 | -0.5107 | Yes | ||
| 126 | PFKFB2 | 3940538 7100059 | 16699 | -0.777 | -0.5157 | Yes | ||
| 127 | NGFRAP1 | 6770195 | 16785 | -0.825 | -0.5098 | Yes | ||
| 128 | GATA3 | 6130068 | 16831 | -0.855 | -0.5015 | Yes | ||
| 129 | EIF4G2 | 3800575 6860184 | 16927 | -0.918 | -0.4950 | Yes | ||
| 130 | TFRC | 4050551 | 16994 | -0.963 | -0.4864 | Yes | ||
| 131 | CD28 | 1400739 4210093 | 17038 | -0.983 | -0.4763 | Yes | ||
| 132 | EIF5 | 6380750 | 17216 | -1.073 | -0.4723 | Yes | ||
| 133 | PPP1R16B | 3520300 | 17237 | -1.091 | -0.4596 | Yes | ||
| 134 | UBE2D2 | 4590671 6510446 | 17253 | -1.102 | -0.4464 | Yes | ||
| 135 | HNRPU | 6520736 | 17265 | -1.114 | -0.4329 | Yes | ||
| 136 | SLC23A2 | 6660722 | 17562 | -1.380 | -0.4315 | Yes | ||
| 137 | CCNE2 | 3120537 | 17567 | -1.388 | -0.4141 | Yes | ||
| 138 | PKNOX1 | 4210452 | 17573 | -1.390 | -0.3968 | Yes | ||
| 139 | GSPT1 | 5420050 | 17593 | -1.408 | -0.3800 | Yes | ||
| 140 | ACBD3 | 2760170 | 17738 | -1.570 | -0.3680 | Yes | ||
| 141 | PHTF2 | 6590053 | 17821 | -1.656 | -0.3514 | Yes | ||
| 142 | ITSN2 | 780484 | 17846 | -1.677 | -0.3315 | Yes | ||
| 143 | GCNT2 | 6400162 2370609 2760309 4540465 6100280 | 17908 | -1.741 | -0.3128 | Yes | ||
| 144 | ZFX | 5900400 | 17912 | -1.744 | -0.2909 | Yes | ||
| 145 | UBE2D3 | 3190452 | 18074 | -1.915 | -0.2754 | Yes | ||
| 146 | MTPN | 3940167 | 18216 | -2.138 | -0.2559 | Yes | ||
| 147 | PABPN1 | 460519 | 18274 | -2.228 | -0.2308 | Yes | ||
| 148 | KPNA1 | 5270324 | 18312 | -2.308 | -0.2036 | Yes | ||
| 149 | HAT1 | 3710082 | 18315 | -2.321 | -0.1744 | Yes | ||
| 150 | MAPK6 | 6760520 | 18324 | -2.346 | -0.1451 | Yes | ||
| 151 | SIN3A | 2190121 | 18425 | -2.561 | -0.1181 | Yes | ||
| 152 | ARID1A | 2630022 1690551 4810110 | 18538 | -2.995 | -0.0863 | Yes | ||
| 153 | APPBP2 | 5130215 | 18579 | -3.469 | -0.0445 | Yes | ||
| 154 | MARCKS | 7040450 | 18595 | -3.674 | 0.0011 | Yes |

