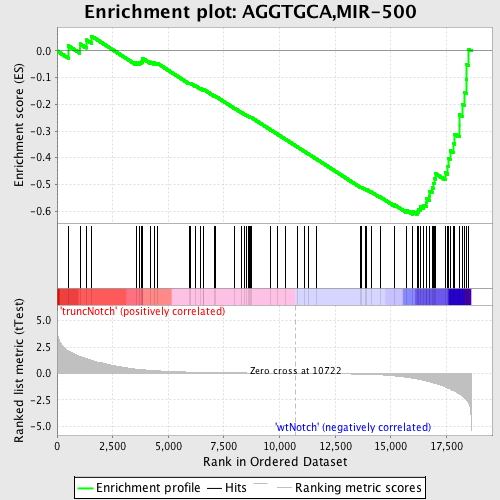
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | AGGTGCA,MIR-500 |
| Enrichment Score (ES) | -0.6127702 |
| Normalized Enrichment Score (NES) | -1.6857612 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.016911611 |
| FWER p-Value | 0.142 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | EGLN2 | 540086 | 497 | 2.139 | 0.0200 | No | ||
| 2 | SLC25A26 | 4210577 | 1028 | 1.593 | 0.0262 | No | ||
| 3 | NOLA2 | 4060167 | 1312 | 1.388 | 0.0413 | No | ||
| 4 | HNRPA0 | 2680048 | 1551 | 1.239 | 0.0556 | No | ||
| 5 | FBXW7 | 4210338 7050280 | 3558 | 0.405 | -0.0437 | No | ||
| 6 | RNF40 | 3610397 | 3712 | 0.371 | -0.0439 | No | ||
| 7 | POFUT1 | 1570458 | 3790 | 0.354 | -0.0403 | No | ||
| 8 | MYNN | 1980022 4210358 | 3839 | 0.347 | -0.0353 | No | ||
| 9 | MATR3 | 1940170 5340278 | 3846 | 0.346 | -0.0280 | No | ||
| 10 | RNF38 | 1940746 3140133 3840601 | 4208 | 0.283 | -0.0413 | No | ||
| 11 | SP8 | 4060576 | 4366 | 0.264 | -0.0440 | No | ||
| 12 | RAB21 | 5570070 | 4511 | 0.242 | -0.0465 | No | ||
| 13 | PLXDC2 | 6400358 | 5928 | 0.123 | -0.1201 | No | ||
| 14 | CELSR3 | 5270731 7100082 | 5981 | 0.121 | -0.1203 | No | ||
| 15 | OSBPL10 | 1980184 | 6215 | 0.108 | -0.1305 | No | ||
| 16 | CHRD | 3140368 | 6444 | 0.097 | -0.1407 | No | ||
| 17 | SYT8 | 4280551 | 6569 | 0.092 | -0.1453 | No | ||
| 18 | LBP | 6860019 | 6597 | 0.091 | -0.1448 | No | ||
| 19 | TSSK2 | 5220324 | 7056 | 0.075 | -0.1679 | No | ||
| 20 | RAI2 | 6770333 | 7132 | 0.072 | -0.1703 | No | ||
| 21 | HLF | 2370113 | 7990 | 0.050 | -0.2154 | No | ||
| 22 | LRRTM3 | 4210021 | 8308 | 0.042 | -0.2316 | No | ||
| 23 | GPM6A | 1660044 2750152 | 8418 | 0.040 | -0.2366 | No | ||
| 24 | PHOX2B | 5270075 | 8517 | 0.038 | -0.2411 | No | ||
| 25 | FIGN | 7000601 | 8589 | 0.037 | -0.2441 | No | ||
| 26 | PRKCE | 5700053 | 8661 | 0.035 | -0.2471 | No | ||
| 27 | GABRB3 | 4150164 | 8713 | 0.034 | -0.2491 | No | ||
| 28 | BTBD11 | 6650154 | 8730 | 0.034 | -0.2493 | No | ||
| 29 | SHPRH | 3990458 | 9613 | 0.018 | -0.2964 | No | ||
| 30 | ESRRG | 4010181 | 9926 | 0.013 | -0.3130 | No | ||
| 31 | SPG3A | 2760091 3120170 | 10276 | 0.007 | -0.3316 | No | ||
| 32 | DRD1 | 430025 | 10806 | -0.001 | -0.3601 | No | ||
| 33 | TBX2 | 1990563 | 11105 | -0.006 | -0.3760 | No | ||
| 34 | HSPA14 | 4610487 | 11286 | -0.009 | -0.3855 | No | ||
| 35 | CACNB1 | 2940427 3710487 | 11679 | -0.016 | -0.4063 | No | ||
| 36 | DDX3Y | 1580278 4200519 | 13648 | -0.075 | -0.5108 | No | ||
| 37 | HIPK1 | 110193 | 13682 | -0.076 | -0.5109 | No | ||
| 38 | E2F3 | 50162 460180 | 13868 | -0.086 | -0.5190 | No | ||
| 39 | RTN4RL1 | 2630368 | 13919 | -0.089 | -0.5197 | No | ||
| 40 | SORCS1 | 60411 5890373 | 14120 | -0.102 | -0.5283 | No | ||
| 41 | KPNA3 | 7040088 | 14523 | -0.137 | -0.5470 | No | ||
| 42 | NRF1 | 2650195 | 15163 | -0.233 | -0.5763 | No | ||
| 43 | CREB1 | 1500717 2230358 3610600 6550601 | 15718 | -0.374 | -0.5980 | No | ||
| 44 | SOCS2 | 4760692 | 15983 | -0.461 | -0.6021 | Yes | ||
| 45 | CIT | 2370601 | 16181 | -0.528 | -0.6012 | Yes | ||
| 46 | ING4 | 510632 2340619 2850176 5290706 | 16248 | -0.554 | -0.5927 | Yes | ||
| 47 | ABCC4 | 130309 | 16325 | -0.586 | -0.5840 | Yes | ||
| 48 | THAP11 | 6940025 | 16478 | -0.651 | -0.5779 | Yes | ||
| 49 | PURB | 5360138 | 16587 | -0.701 | -0.5684 | Yes | ||
| 50 | KLF9 | 3840022 | 16607 | -0.722 | -0.5537 | Yes | ||
| 51 | NFAT5 | 2510411 5890195 6550152 | 16745 | -0.804 | -0.5435 | Yes | ||
| 52 | CAMK4 | 1690091 | 16754 | -0.806 | -0.5263 | Yes | ||
| 53 | SEC24C | 6980707 | 16855 | -0.869 | -0.5127 | Yes | ||
| 54 | PPP3CC | 2450139 | 16907 | -0.903 | -0.4957 | Yes | ||
| 55 | RBMS1 | 6400014 | 16951 | -0.929 | -0.4777 | Yes | ||
| 56 | CUTL1 | 1570113 1770091 1850273 2100286 5340605 | 17006 | -0.971 | -0.4594 | Yes | ||
| 57 | KBTBD2 | 2320014 | 17441 | -1.261 | -0.4552 | Yes | ||
| 58 | TIMM8A | 110279 | 17566 | -1.388 | -0.4316 | Yes | ||
| 59 | SLC4A7 | 2370056 | 17589 | -1.404 | -0.4021 | Yes | ||
| 60 | DDX3X | 2190020 | 17667 | -1.484 | -0.3738 | Yes | ||
| 61 | PPP4R1 | 5670088 6420402 | 17808 | -1.641 | -0.3454 | Yes | ||
| 62 | PPP2R5E | 70671 2120056 | 17878 | -1.710 | -0.3118 | Yes | ||
| 63 | NCOA1 | 3610438 | 18073 | -1.911 | -0.2804 | Yes | ||
| 64 | UBE2D3 | 3190452 | 18074 | -1.915 | -0.2386 | Yes | ||
| 65 | IVNS1ABP | 4760601 6520113 | 18205 | -2.117 | -0.1993 | Yes | ||
| 66 | RAB14 | 6860139 | 18296 | -2.272 | -0.1545 | Yes | ||
| 67 | APRIN | 2810050 | 18392 | -2.494 | -0.1051 | Yes | ||
| 68 | DCTN4 | 3390010 5550170 | 18410 | -2.527 | -0.0508 | Yes | ||
| 69 | OGT | 2360131 4610333 | 18507 | -2.829 | 0.0059 | Yes |

