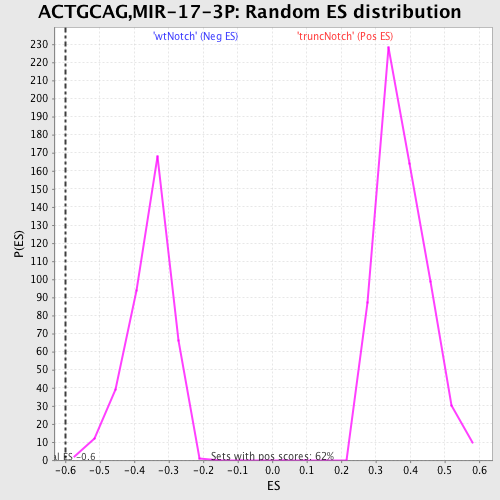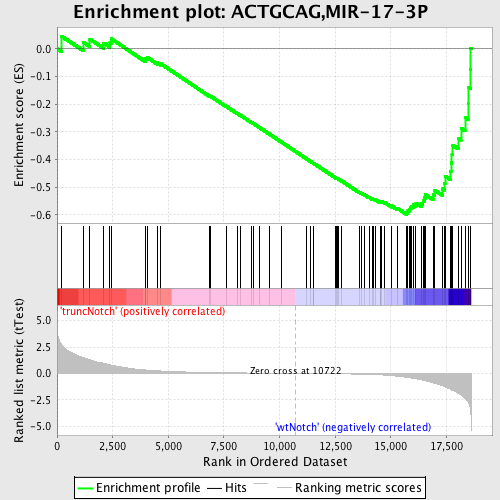
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | ACTGCAG,MIR-17-3P |
| Enrichment Score (ES) | -0.59869045 |
| Normalized Enrichment Score (NES) | -1.6810734 |
| Nominal p-value | 0.002617801 |
| FDR q-value | 0.016509522 |
| FWER p-Value | 0.155 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CAMK2D | 2370685 | 198 | 2.728 | 0.0460 | No | ||
| 2 | ARID2 | 110647 5390435 | 1184 | 1.479 | 0.0236 | No | ||
| 3 | ANKRD50 | 870543 | 1472 | 1.290 | 0.0350 | No | ||
| 4 | RASGRP4 | 540102 | 2101 | 0.945 | 0.0208 | No | ||
| 5 | VGLL4 | 6860463 | 2375 | 0.818 | 0.0231 | No | ||
| 6 | PSCD1 | 110113 2470592 | 2437 | 0.787 | 0.0361 | No | ||
| 7 | ARMC8 | 520408 940132 4060368 4210102 | 3972 | 0.325 | -0.0398 | No | ||
| 8 | UHMK1 | 3850670 6450064 | 3977 | 0.324 | -0.0333 | No | ||
| 9 | ACTR1A | 5860070 | 4070 | 0.308 | -0.0318 | No | ||
| 10 | RAB21 | 5570070 | 4511 | 0.242 | -0.0505 | No | ||
| 11 | ACACA | 2490612 2680369 | 4667 | 0.226 | -0.0542 | No | ||
| 12 | TM9SF4 | 450538 1050093 4210112 | 6848 | 0.082 | -0.1700 | No | ||
| 13 | FZD4 | 1580520 | 6907 | 0.079 | -0.1715 | No | ||
| 14 | RGAG4 | 1170167 | 7594 | 0.059 | -0.2073 | No | ||
| 15 | TCERG1L | 3360707 | 8108 | 0.047 | -0.2339 | No | ||
| 16 | MEIS1 | 1400575 | 8223 | 0.044 | -0.2392 | No | ||
| 17 | COL12A1 | 1990025 3780086 | 8749 | 0.033 | -0.2668 | No | ||
| 18 | HIF1A | 5670605 | 8830 | 0.032 | -0.2704 | No | ||
| 19 | ZFR | 5910647 | 9092 | 0.027 | -0.2839 | No | ||
| 20 | ITSN1 | 2640577 | 9525 | 0.020 | -0.3068 | No | ||
| 21 | ZFHX4 | 6110167 | 10096 | 0.010 | -0.3373 | No | ||
| 22 | KCNMA1 | 1450402 | 11220 | -0.008 | -0.3977 | No | ||
| 23 | SORCS2 | 6350520 | 11377 | -0.011 | -0.4059 | No | ||
| 24 | MYO1D | 6760292 | 11517 | -0.013 | -0.4131 | No | ||
| 25 | ZIC3 | 380020 | 11532 | -0.013 | -0.4136 | No | ||
| 26 | ZNF462 | 6900100 | 12530 | -0.035 | -0.4667 | No | ||
| 27 | NEUROG1 | 380341 | 12565 | -0.035 | -0.4678 | No | ||
| 28 | NF2 | 4150735 6450139 | 12588 | -0.036 | -0.4682 | No | ||
| 29 | SOX11 | 610279 | 12668 | -0.038 | -0.4717 | No | ||
| 30 | LIN28 | 6590672 | 12802 | -0.042 | -0.4779 | No | ||
| 31 | USP33 | 780047 | 13574 | -0.071 | -0.5180 | No | ||
| 32 | HMGA2 | 2940121 3390647 5130279 6400136 | 13670 | -0.076 | -0.5216 | No | ||
| 33 | NTRK2 | 6220463 | 13810 | -0.083 | -0.5274 | No | ||
| 34 | PCTK2 | 5700673 | 14026 | -0.096 | -0.5370 | No | ||
| 35 | FSTL5 | 770280 | 14169 | -0.106 | -0.5424 | No | ||
| 36 | SPRED2 | 510138 540195 | 14226 | -0.111 | -0.5431 | No | ||
| 37 | OLFM1 | 3360161 5570066 | 14298 | -0.117 | -0.5445 | No | ||
| 38 | KPNA3 | 7040088 | 14523 | -0.137 | -0.5538 | No | ||
| 39 | DDX5 | 940021 2630338 3440148 | 14526 | -0.137 | -0.5510 | No | ||
| 40 | HDAC8 | 3440504 | 14590 | -0.144 | -0.5514 | No | ||
| 41 | BCORL1 | 1660008 | 14721 | -0.160 | -0.5551 | No | ||
| 42 | RAB6A | 3840463 4780731 | 15031 | -0.206 | -0.5675 | No | ||
| 43 | MYST2 | 4540494 | 15299 | -0.262 | -0.5764 | No | ||
| 44 | INPP5A | 5910195 | 15713 | -0.372 | -0.5910 | Yes | ||
| 45 | RNF111 | 2970072 3140112 5130647 | 15769 | -0.389 | -0.5858 | Yes | ||
| 46 | RAB8B | 510692 1980017 3130136 4570739 | 15822 | -0.409 | -0.5801 | Yes | ||
| 47 | GTF2H1 | 2570746 2650148 | 15865 | -0.423 | -0.5736 | Yes | ||
| 48 | NRD1 | 5900017 | 15942 | -0.448 | -0.5684 | Yes | ||
| 49 | NR3C1 | 630563 | 16015 | -0.470 | -0.5625 | Yes | ||
| 50 | DDX6 | 2630025 6770408 | 16128 | -0.510 | -0.5579 | Yes | ||
| 51 | FOXP1 | 6510433 | 16376 | -0.605 | -0.5587 | Yes | ||
| 52 | BTF3 | 6940279 | 16446 | -0.636 | -0.5492 | Yes | ||
| 53 | PPP1R10 | 110136 3170725 3610484 | 16503 | -0.666 | -0.5384 | Yes | ||
| 54 | CNOT4 | 2320242 6760706 | 16547 | -0.685 | -0.5264 | Yes | ||
| 55 | MYOHD1 | 1410520 | 16906 | -0.903 | -0.5270 | Yes | ||
| 56 | PARP6 | 3290332 4760167 6900181 | 16978 | -0.950 | -0.5111 | Yes | ||
| 57 | KHDRBS1 | 1240403 6040040 | 17319 | -1.152 | -0.5054 | Yes | ||
| 58 | RAB11A | 5360079 | 17419 | -1.242 | -0.4850 | Yes | ||
| 59 | KBTBD2 | 2320014 | 17441 | -1.261 | -0.4599 | Yes | ||
| 60 | YTHDF2 | 4060332 4610279 | 17690 | -1.501 | -0.4421 | Yes | ||
| 61 | FKBP5 | 2190048 | 17709 | -1.535 | -0.4111 | Yes | ||
| 62 | YWHAG | 3780341 | 17748 | -1.581 | -0.3803 | Yes | ||
| 63 | MAX | 5220021 | 17789 | -1.621 | -0.3488 | Yes | ||
| 64 | RAP2C | 1690132 | 18028 | -1.868 | -0.3228 | Yes | ||
| 65 | FGFR1OP | 6510300 | 18156 | -2.046 | -0.2871 | Yes | ||
| 66 | EIF4G1 | 4070446 | 18338 | -2.375 | -0.2475 | Yes | ||
| 67 | RHOT1 | 6760692 | 18504 | -2.826 | -0.1977 | Yes | ||
| 68 | OGT | 2360131 4610333 | 18507 | -2.829 | -0.1390 | Yes | ||
| 69 | PPP2CA | 3990113 | 18569 | -3.290 | -0.0739 | Yes | ||
| 70 | MARCKS | 7040450 | 18595 | -3.674 | 0.0011 | Yes |