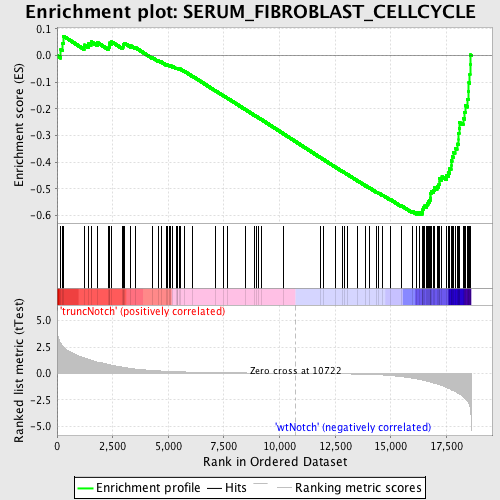
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | SERUM_FIBROBLAST_CELLCYCLE |
| Enrichment Score (ES) | -0.5961463 |
| Normalized Enrichment Score (NES) | -1.7531214 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.042854153 |
| FWER p-Value | 0.39 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MBOAT1 | 4010309 | 168 | 2.809 | 0.0227 | No | ||
| 2 | SDC1 | 3440471 6350408 | 255 | 2.549 | 0.0470 | No | ||
| 3 | TYMS | 940450 1940068 3710008 5570546 | 288 | 2.473 | 0.0733 | No | ||
| 4 | TIMP1 | 1010326 | 1212 | 1.464 | 0.0400 | No | ||
| 5 | H2AFX | 3520082 | 1412 | 1.332 | 0.0443 | No | ||
| 6 | KIFC1 | 2570332 2480725 | 1525 | 1.254 | 0.0525 | No | ||
| 7 | RANGAP1 | 2320593 6650601 | 1814 | 1.071 | 0.0490 | No | ||
| 8 | TUBB | 730670 3390037 | 2327 | 0.844 | 0.0310 | No | ||
| 9 | ASF1B | 6590706 | 2342 | 0.835 | 0.0397 | No | ||
| 10 | PPIH | 5910403 | 2370 | 0.824 | 0.0475 | No | ||
| 11 | ATAD2 | 870242 2260687 | 2457 | 0.780 | 0.0517 | No | ||
| 12 | SGCD | 2940685 5080753 | 2946 | 0.584 | 0.0320 | No | ||
| 13 | PAQR4 | 730736 | 2964 | 0.576 | 0.0376 | No | ||
| 14 | CIITA | 2650242 | 2990 | 0.568 | 0.0427 | No | ||
| 15 | MAPK13 | 6760215 | 3027 | 0.553 | 0.0470 | No | ||
| 16 | EXO1 | 3610408 6040008 | 3314 | 0.460 | 0.0367 | No | ||
| 17 | MCM5 | 2680647 | 3531 | 0.411 | 0.0297 | No | ||
| 18 | GTSE1 | 3840369 4280273 | 4273 | 0.274 | -0.0072 | No | ||
| 19 | FOXM1 | 6650402 6980091 | 4550 | 0.238 | -0.0194 | No | ||
| 20 | USP1 | 2760541 5690438 6220673 | 4671 | 0.225 | -0.0233 | No | ||
| 21 | ITGB3 | 5270463 | 4936 | 0.198 | -0.0354 | No | ||
| 22 | RECQL4 | 540551 | 4966 | 0.195 | -0.0347 | No | ||
| 23 | ANLN | 1780113 2470537 6840047 | 5049 | 0.187 | -0.0370 | No | ||
| 24 | MLLT6 | 3870168 | 5112 | 0.182 | -0.0383 | No | ||
| 25 | UBE2C | 6130017 | 5204 | 0.173 | -0.0413 | No | ||
| 26 | FANCG | 4010471 | 5351 | 0.161 | -0.0473 | No | ||
| 27 | PLK1 | 1780369 2640121 | 5389 | 0.159 | -0.0475 | No | ||
| 28 | CDCA5 | 5670131 | 5416 | 0.156 | -0.0472 | No | ||
| 29 | HN1 | 3360156 | 5509 | 0.150 | -0.0504 | No | ||
| 30 | HMMR | 5720315 6380168 | 5531 | 0.148 | -0.0499 | No | ||
| 31 | MCM8 | 6130743 | 5717 | 0.135 | -0.0584 | No | ||
| 32 | HIST1H2AC | 3130292 | 6094 | 0.114 | -0.0774 | No | ||
| 33 | MET | 2690348 | 7105 | 0.073 | -0.1311 | No | ||
| 34 | EFHC1 | 2570288 | 7481 | 0.062 | -0.1506 | No | ||
| 35 | FANCA | 6130070 | 7650 | 0.058 | -0.1591 | No | ||
| 36 | CDKN3 | 110520 | 8482 | 0.039 | -0.2035 | No | ||
| 37 | ABCC5 | 2100600 5050692 | 8884 | 0.031 | -0.2248 | No | ||
| 38 | HELLS | 4560086 4810025 | 8968 | 0.029 | -0.2290 | No | ||
| 39 | CKS2 | 1410156 | 9063 | 0.028 | -0.2337 | No | ||
| 40 | ADAMTS1 | 3140286 5860463 | 9190 | 0.025 | -0.2402 | No | ||
| 41 | BUB1 | 5390270 | 10159 | 0.009 | -0.2924 | No | ||
| 42 | CCNF | 6370288 | 11830 | -0.019 | -0.3824 | No | ||
| 43 | CENPF | 5050088 | 11951 | -0.022 | -0.3886 | No | ||
| 44 | ESCO2 | 1170010 | 12528 | -0.034 | -0.4193 | No | ||
| 45 | CDKL5 | 2450070 | 12835 | -0.043 | -0.4354 | No | ||
| 46 | YWHAH | 1660133 2810053 | 12901 | -0.045 | -0.4384 | No | ||
| 47 | CCNB2 | 6510528 | 13046 | -0.050 | -0.4456 | No | ||
| 48 | DHFR | 6350315 | 13517 | -0.069 | -0.4702 | No | ||
| 49 | PTTG1 | 520463 | 13883 | -0.086 | -0.4889 | No | ||
| 50 | MLF1IP | 3610367 | 14046 | -0.098 | -0.4966 | No | ||
| 51 | CDC25C | 2570673 4760161 6520707 | 14375 | -0.124 | -0.5129 | No | ||
| 52 | CASP3 | 6620397 | 14460 | -0.131 | -0.5159 | No | ||
| 53 | EZH2 | 6130605 6380524 | 14633 | -0.150 | -0.5235 | No | ||
| 54 | AMD1 | 6290128 | 14974 | -0.198 | -0.5396 | No | ||
| 55 | FEN1 | 1770541 | 15480 | -0.302 | -0.5635 | No | ||
| 56 | PRIM2A | 7000692 | 15959 | -0.456 | -0.5842 | No | ||
| 57 | SMC4 | 5910240 | 16168 | -0.524 | -0.5894 | No | ||
| 58 | BARD1 | 3170348 | 16293 | -0.574 | -0.5896 | Yes | ||
| 59 | CDC25A | 3800184 | 16404 | -0.617 | -0.5886 | Yes | ||
| 60 | FBXL20 | 4200064 | 16431 | -0.629 | -0.5829 | Yes | ||
| 61 | CCNA2 | 5290075 | 16440 | -0.634 | -0.5761 | Yes | ||
| 62 | CDC6 | 4570296 5360600 | 16457 | -0.641 | -0.5697 | Yes | ||
| 63 | TRIP13 | 6860341 | 16493 | -0.659 | -0.5642 | Yes | ||
| 64 | DONSON | 4780541 6130010 | 16611 | -0.725 | -0.5623 | Yes | ||
| 65 | MELK | 130022 5220537 | 16632 | -0.736 | -0.5550 | Yes | ||
| 66 | DLG7 | 3120041 | 16674 | -0.763 | -0.5486 | Yes | ||
| 67 | CENPA | 5080154 | 16744 | -0.803 | -0.5432 | Yes | ||
| 68 | TACC3 | 5130592 | 16768 | -0.814 | -0.5353 | Yes | ||
| 69 | MCM6 | 60092 540181 6510110 | 16794 | -0.831 | -0.5272 | Yes | ||
| 70 | SPAG5 | 6200288 6940520 | 16795 | -0.831 | -0.5178 | Yes | ||
| 71 | LMNB1 | 5890292 6020008 | 16813 | -0.842 | -0.5092 | Yes | ||
| 72 | GMNN | 2630148 | 16925 | -0.916 | -0.5048 | Yes | ||
| 73 | KNTC1 | 430079 | 16964 | -0.940 | -0.4962 | Yes | ||
| 74 | PRIM1 | 6420746 | 17077 | -0.999 | -0.4909 | Yes | ||
| 75 | DEPDC1B | 2120458 | 17160 | -1.038 | -0.4836 | Yes | ||
| 76 | RRM2 | 6350059 6940162 | 17182 | -1.046 | -0.4729 | Yes | ||
| 77 | TIPIN | 1570301 2480113 | 17184 | -1.048 | -0.4611 | Yes | ||
| 78 | AURKA | 780537 | 17299 | -1.140 | -0.4543 | Yes | ||
| 79 | CDCA8 | 2340286 6980019 | 17502 | -1.320 | -0.4503 | Yes | ||
| 80 | KIF23 | 5570112 | 17597 | -1.409 | -0.4394 | Yes | ||
| 81 | ILF2 | 2900253 | 17637 | -1.455 | -0.4250 | Yes | ||
| 82 | TOP2A | 360717 | 17707 | -1.531 | -0.4114 | Yes | ||
| 83 | MCM4 | 2760673 5420711 | 17712 | -1.538 | -0.3942 | Yes | ||
| 84 | MPHOSPH1 | 4610427 | 17751 | -1.584 | -0.3783 | Yes | ||
| 85 | PHTF2 | 6590053 | 17821 | -1.656 | -0.3633 | Yes | ||
| 86 | RAD51AP1 | 4050736 6370528 | 17885 | -1.714 | -0.3473 | Yes | ||
| 87 | UHRF1 | 1410273 1660242 | 17976 | -1.817 | -0.3316 | Yes | ||
| 88 | RRM1 | 4150433 | 18035 | -1.872 | -0.3135 | Yes | ||
| 89 | PBK | 5670600 | 18037 | -1.875 | -0.2923 | Yes | ||
| 90 | ANKRD10 | 1240025 | 18068 | -1.906 | -0.2724 | Yes | ||
| 91 | PCNA | 940754 | 18090 | -1.941 | -0.2515 | Yes | ||
| 92 | CKAP2 | 3520692 | 18250 | -2.192 | -0.2353 | Yes | ||
| 93 | TPX2 | 6420324 | 18310 | -2.306 | -0.2124 | Yes | ||
| 94 | MAD2L1 | 4480725 | 18343 | -2.379 | -0.1872 | Yes | ||
| 95 | ANP32E | 6510706 | 18450 | -2.630 | -0.1631 | Yes | ||
| 96 | CDCA7 | 3060097 | 18500 | -2.812 | -0.1339 | Yes | ||
| 97 | LYAR | 7000402 | 18509 | -2.840 | -0.1022 | Yes | ||
| 98 | RFC4 | 3800082 6840142 | 18552 | -3.081 | -0.0696 | Yes | ||
| 99 | NCAPH | 6220435 | 18560 | -3.183 | -0.0339 | Yes | ||
| 100 | WSB1 | 870563 | 18567 | -3.256 | 0.0026 | Yes |

